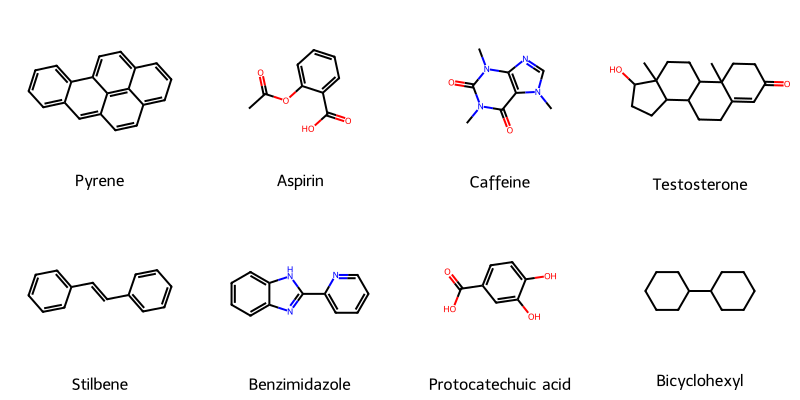
Molecular String Renderer: Robust Visualization Tool
A robust, type-safe Python library for converting chemical strings (SMILES, SELFIES, InChI) into publication-quality …

A robust, type-safe Python library for converting chemical strings (SMILES, SELFIES, InChI) into publication-quality …

GPU-accelerated PyTorch framework for the Müller-Brown potential with JIT compilation, Langevin dynamics, and …
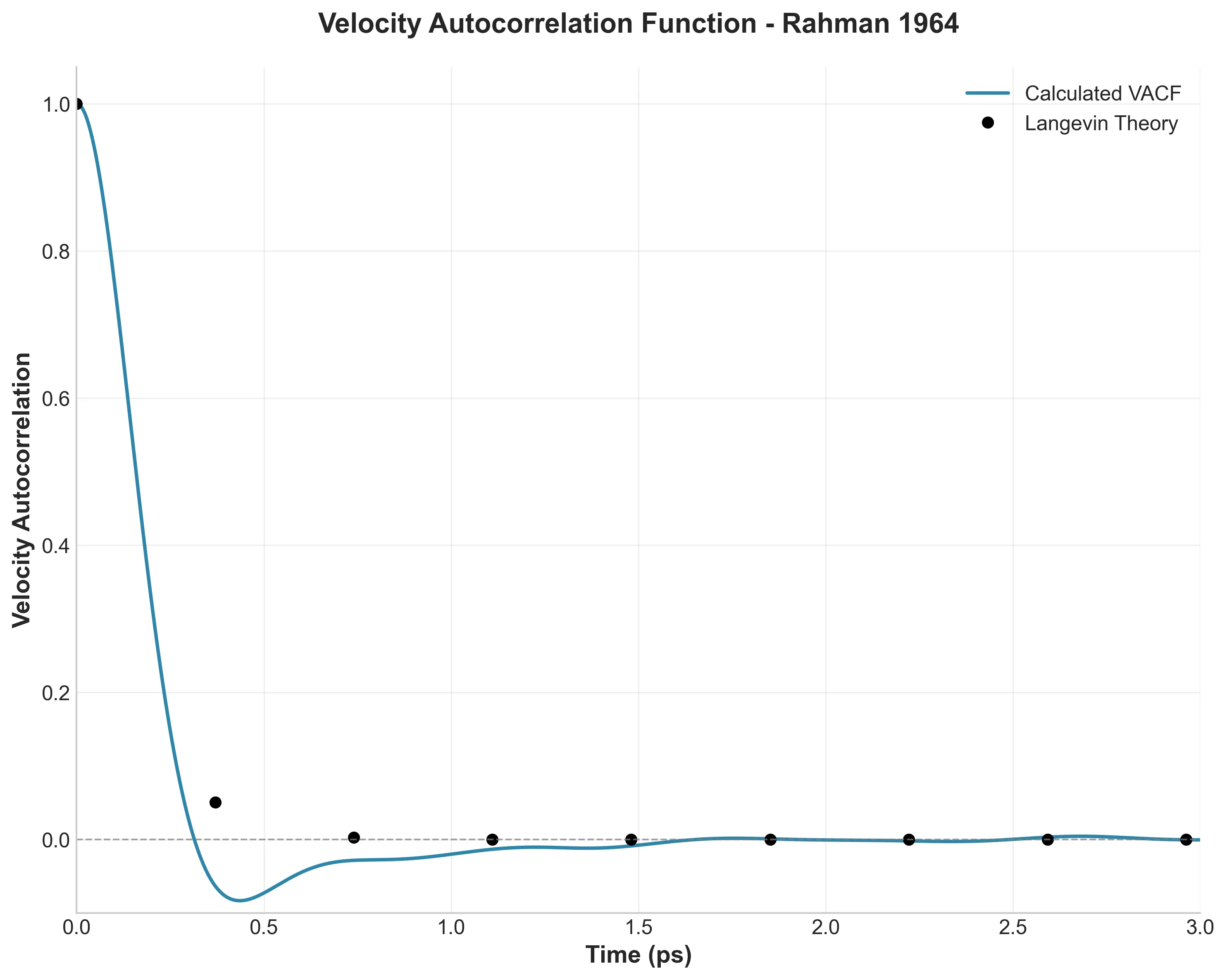
A high-fidelity replication of foundational molecular dynamics using modern software engineering practices: caching, …
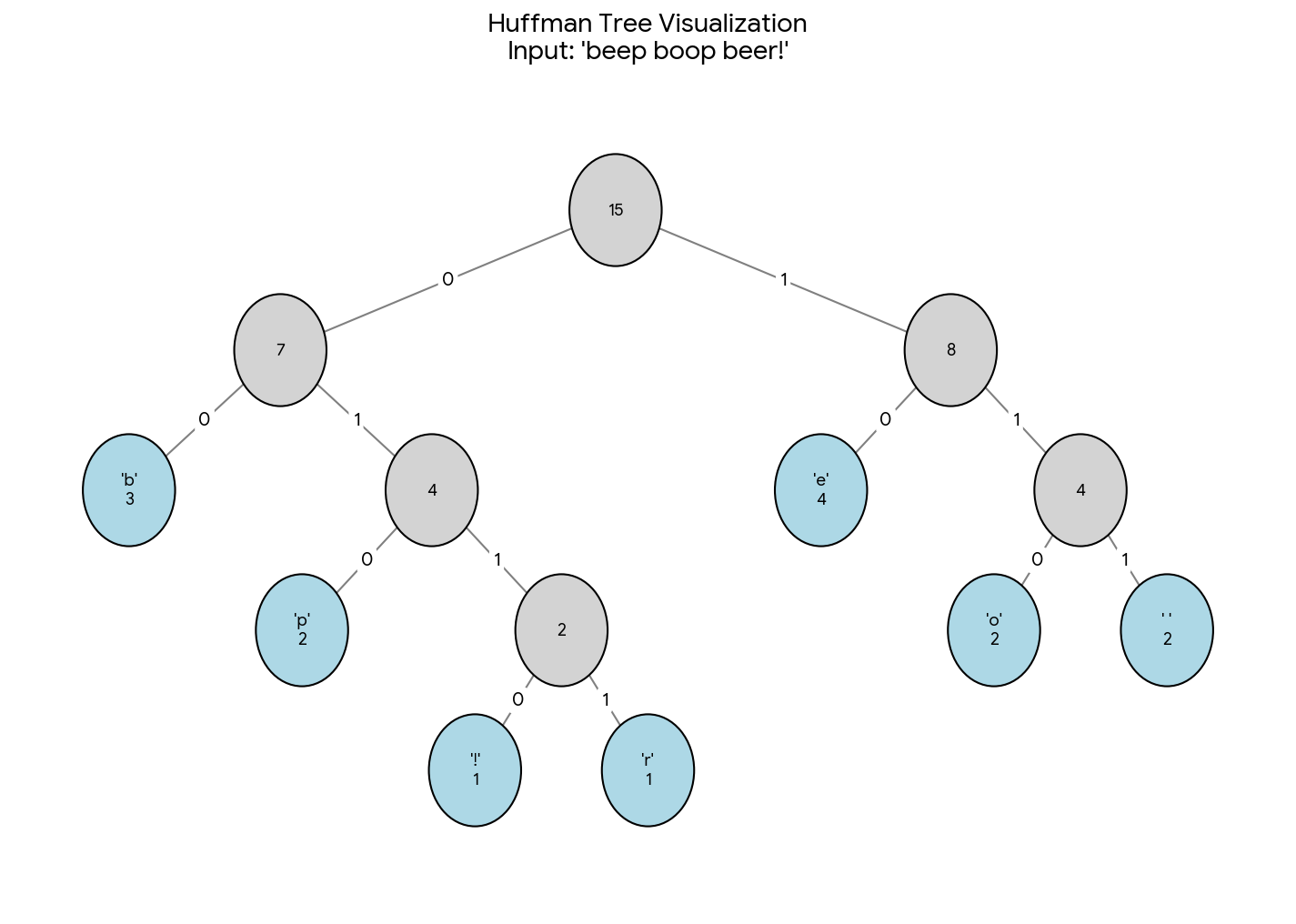
Production-grade Word2Vec in PyTorch with vectorized Hierarchical Softmax, Negative Sampling, and torch.compile support.
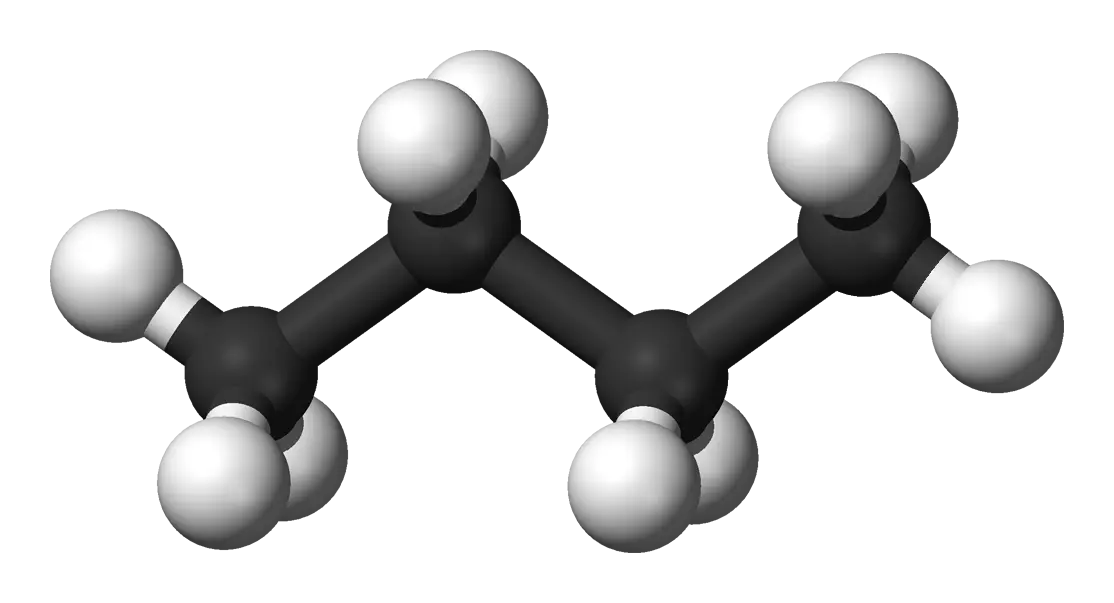
An end-to-end cheminformatics pipeline transforming 1D chemical formulas into 3D conformer datasets using graph …
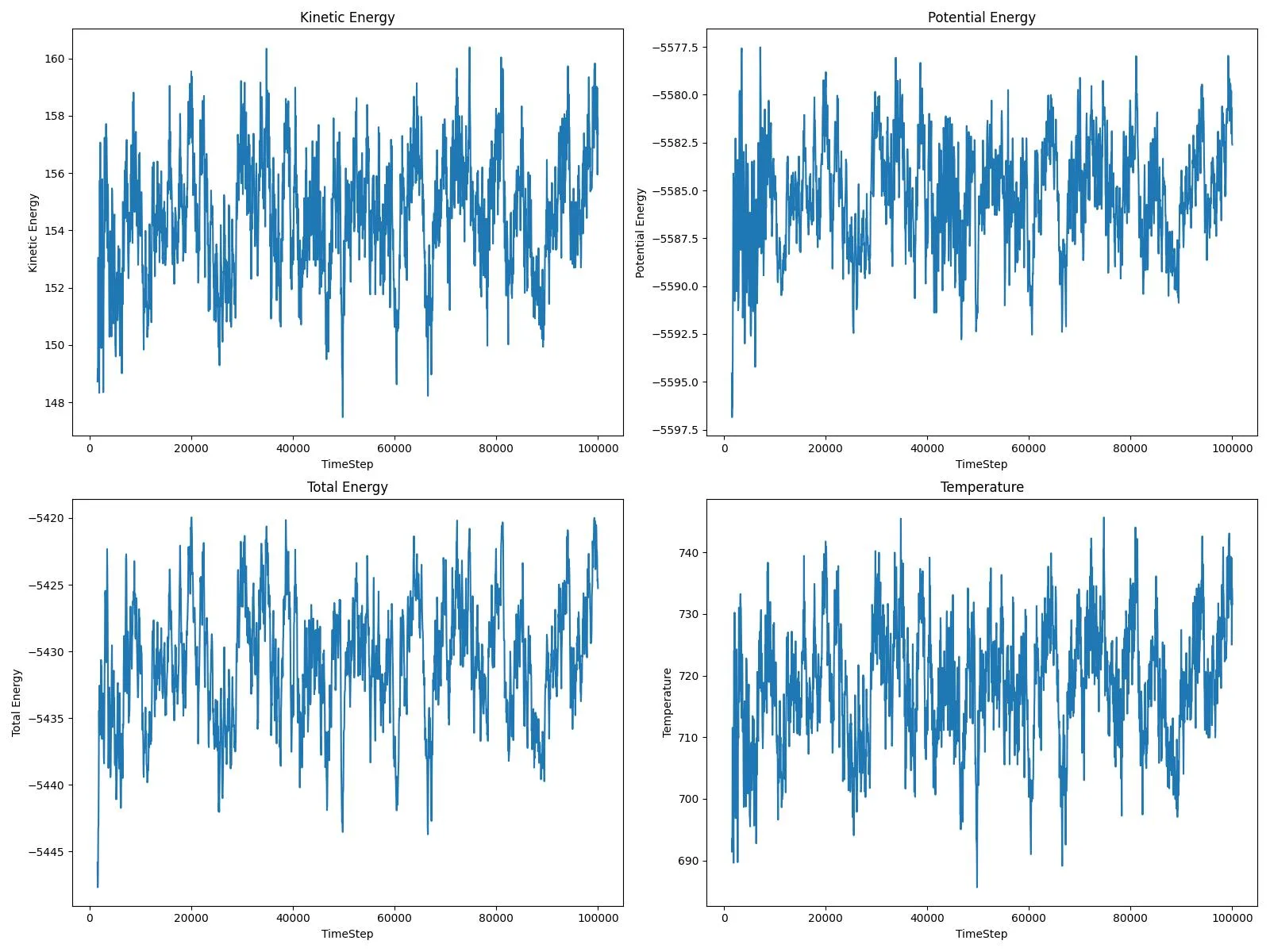
Python-wrapped reference implementation for surface diffusion simulations using LAMMPS and EAM potentials, with …
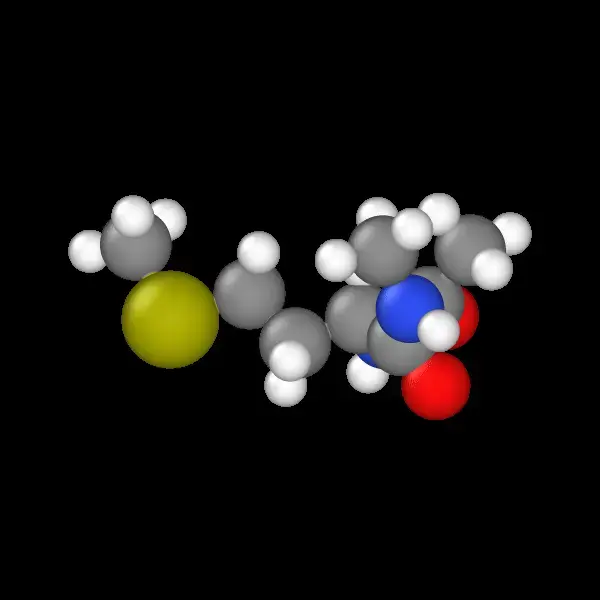
Automated GROMACS pipeline generating high-fidelity MD trajectories with atomic force extraction for Neural Network …
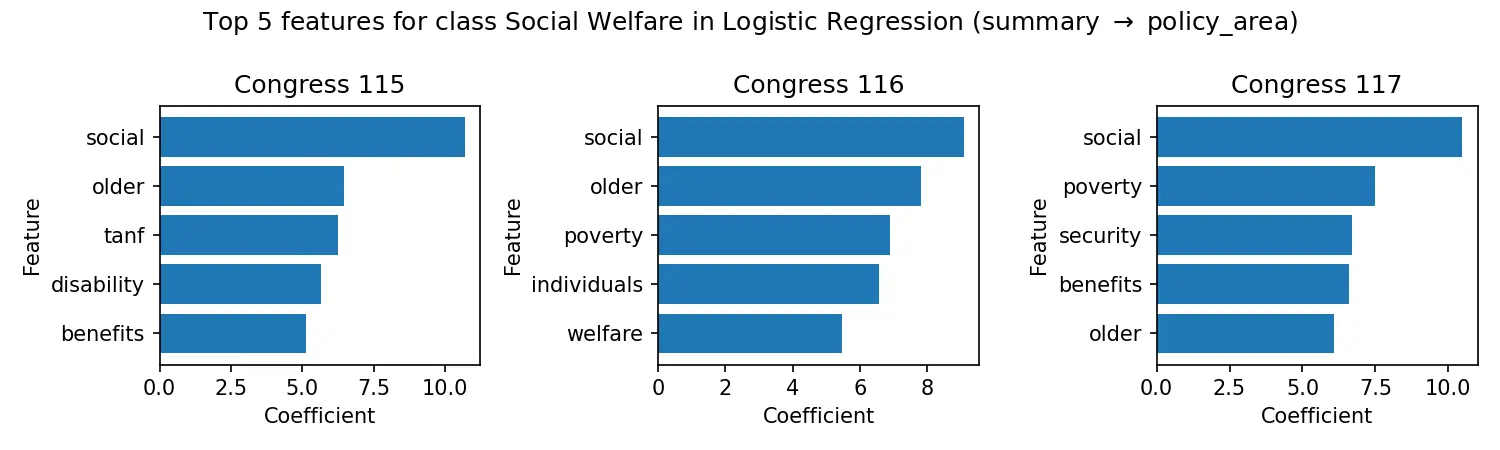
A 47,000+ bill knowledge graph from Congress.gov with sponsor networks and 87% policy classification accuracy.
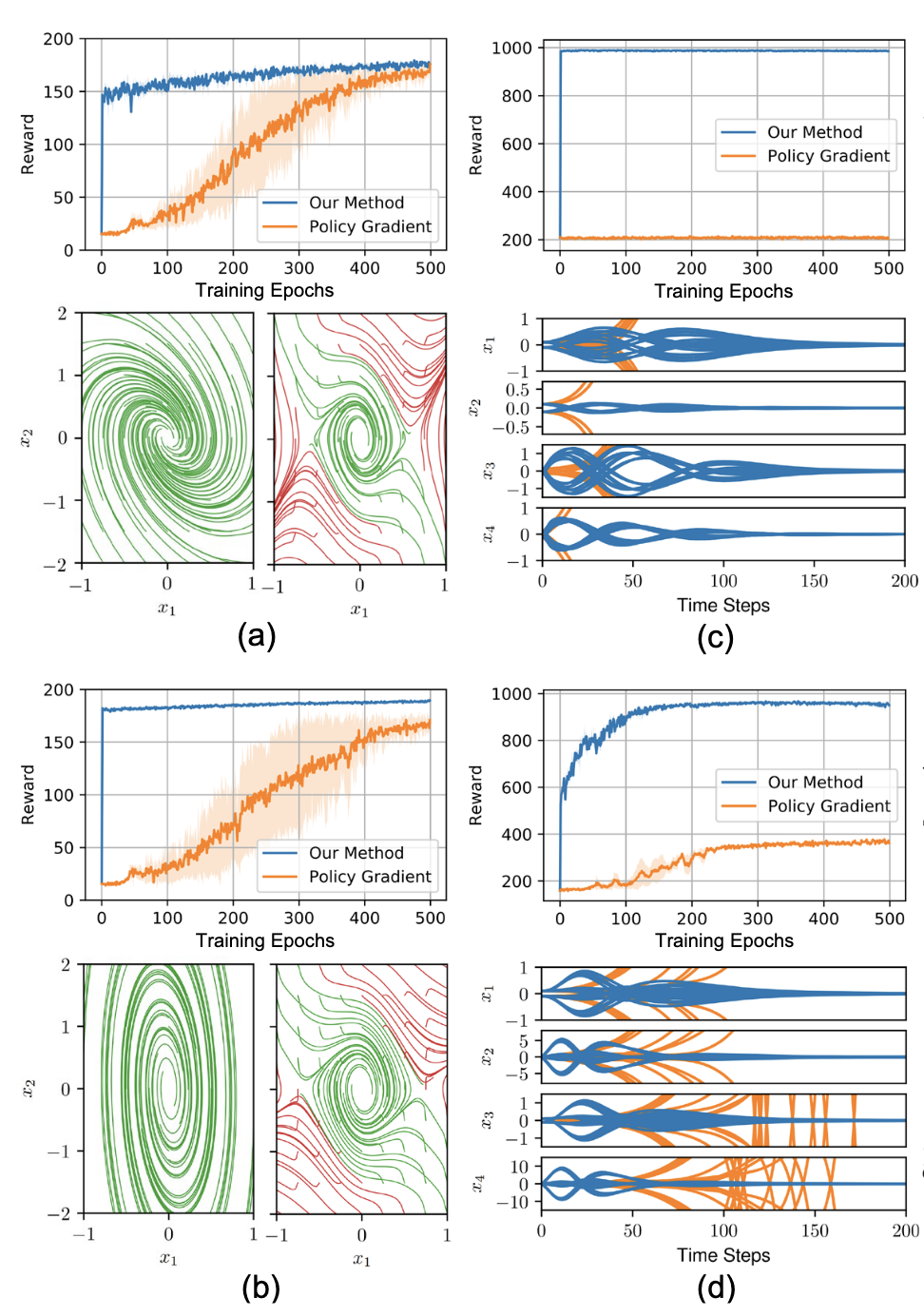
PyTorch IQCRNN enforcing stability guarantees on RNNs via Integral Quadratic Constraints and semidefinite programming.

Undergraduate thesis exploring representation learning for social media text and developing tools for cross-platform …
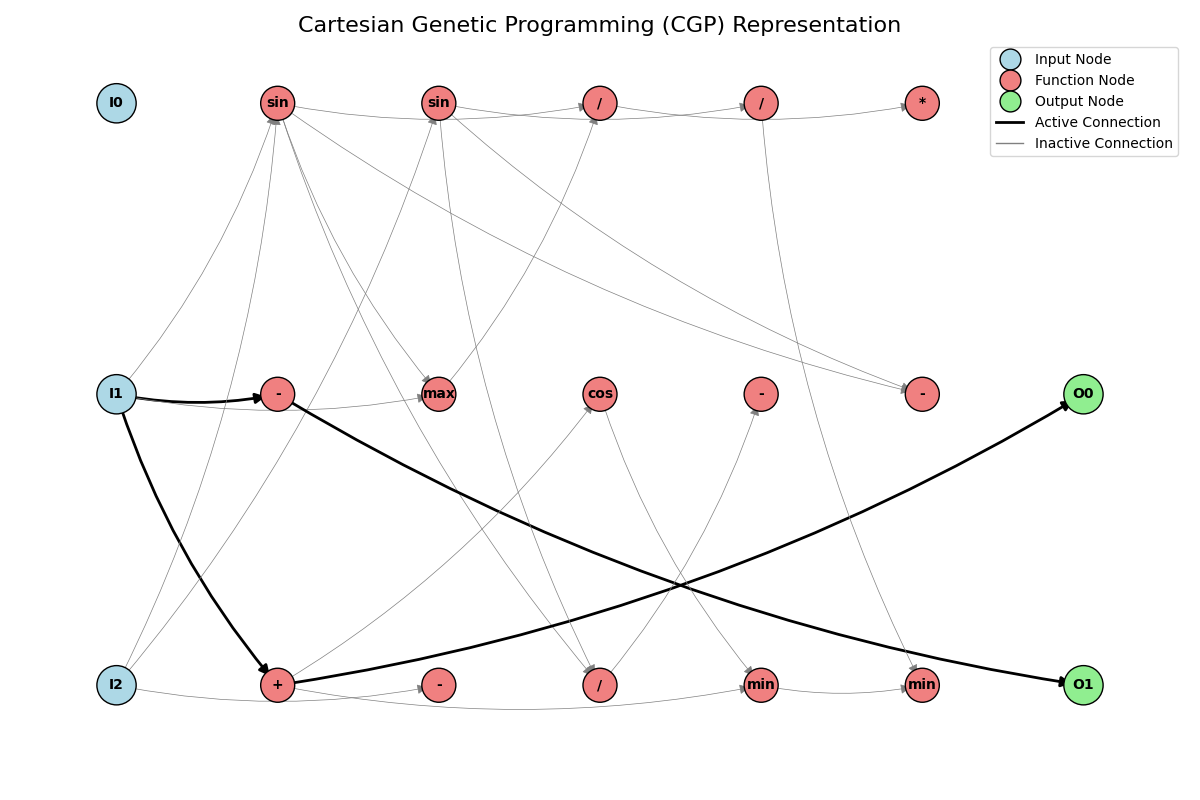
An implementation of Cartesian Genetic Programming (CGP) featuring NEAT-like speciation mechanics and novel crossover …
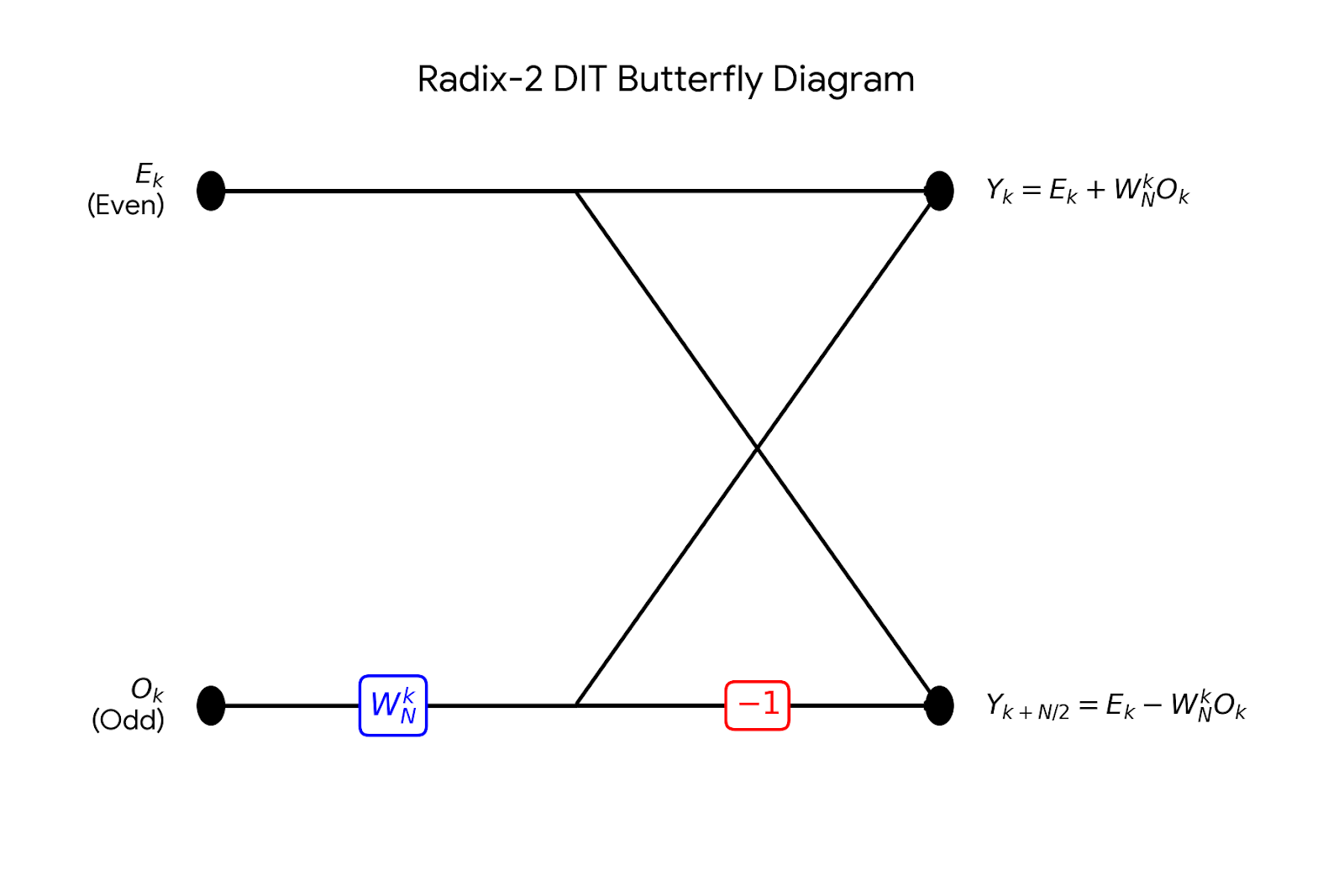
Reverse-engineering the genfft logic to generate optimized C kernels for Fast Fourier Transforms using Haskell …