
ChemBERTa-3: Open Source Training Framework
An open-source framework integrating DeepChem and Ray for training and benchmarking chemical foundation models like …

An open-source framework integrating DeepChem and Ray for training and benchmarking chemical foundation models like …
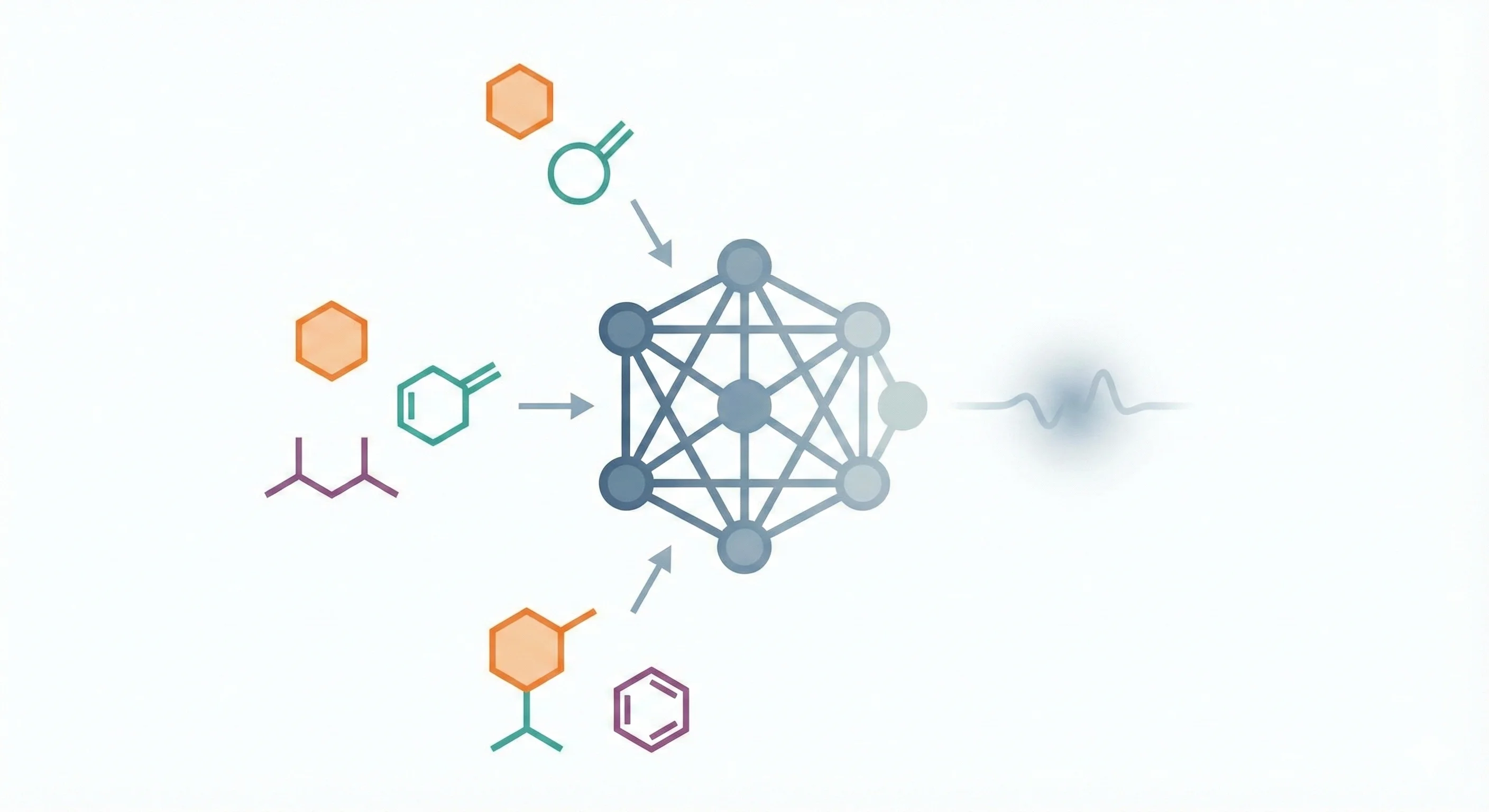
A 14B-parameter chemical reasoning LLM enhanced with atomized functional group knowledge and mix-sourced distillation …

Optimizing transformer pretraining for molecules using MLM vs MTR objectives, scaling to 77M compounds from PubChem for …
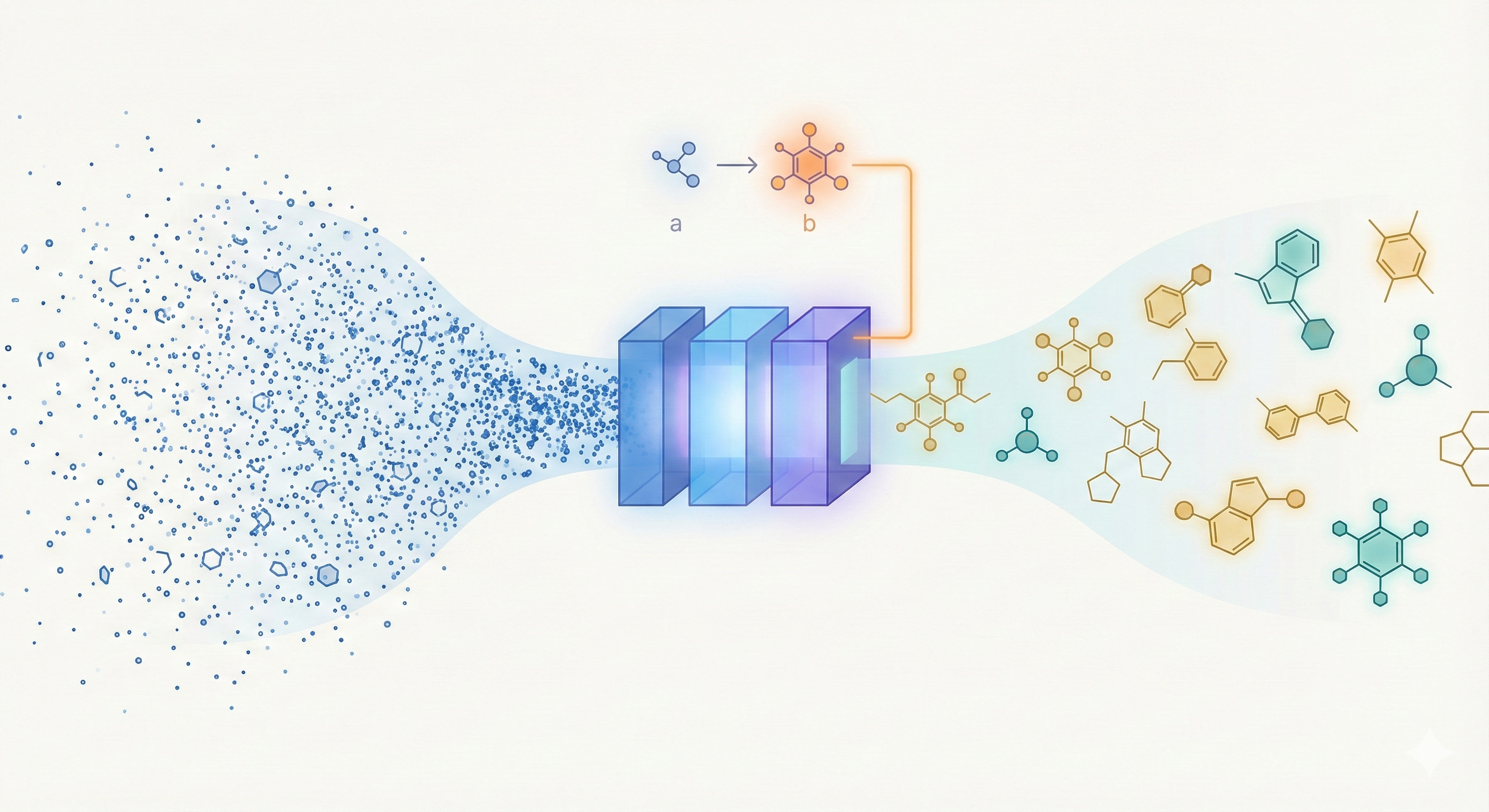
A 46.8M parameter transformer for molecular generation trained on 1.1B SMILES, introducing pair-tuning for efficient …

A systematic evaluation of RoBERTa transformers pretrained on 77M PubChem SMILES for molecular property prediction …
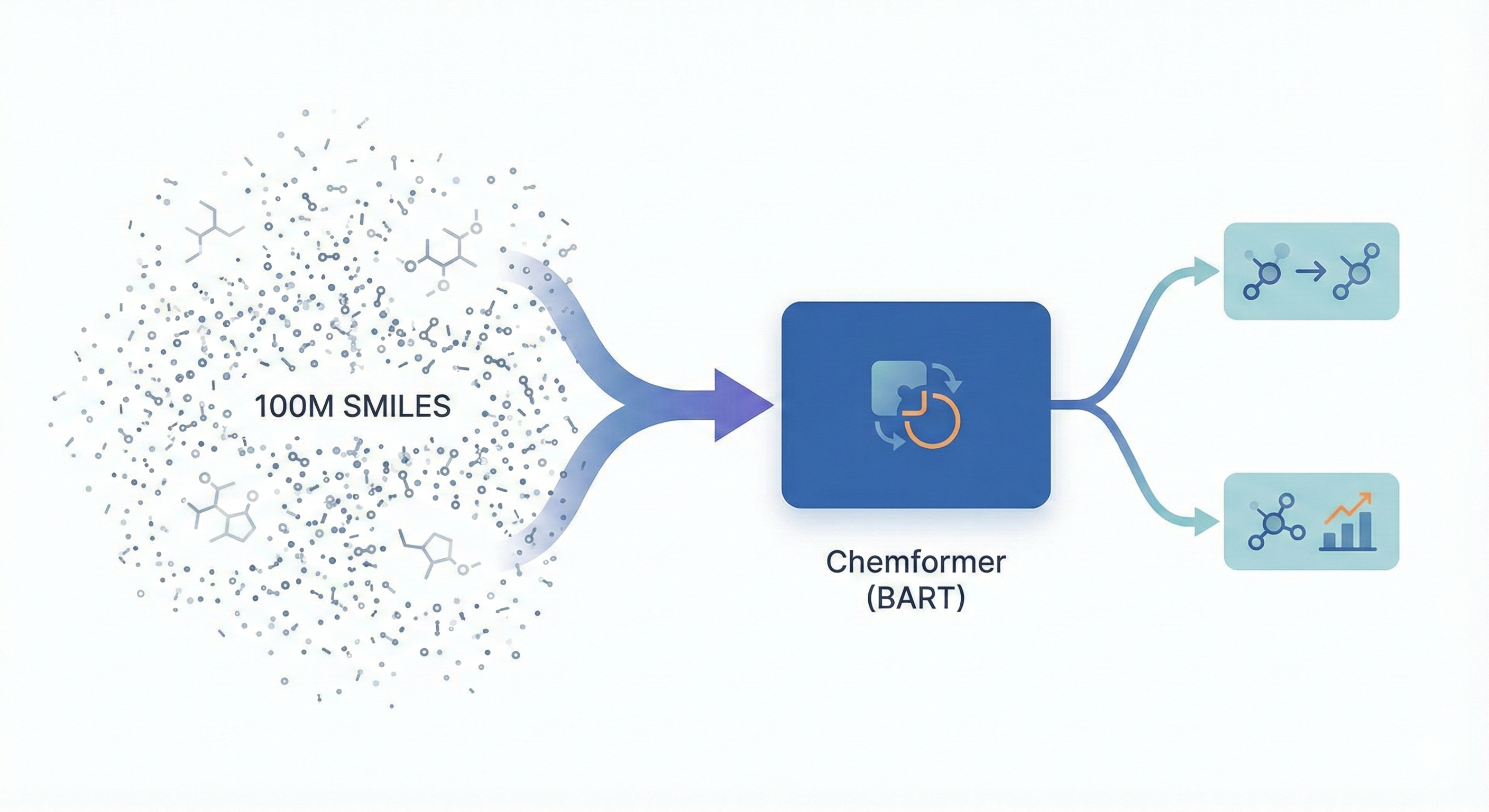
BART-based Transformer pre-trained on 100M molecules using self-supervision to accelerate convergence on chemical …
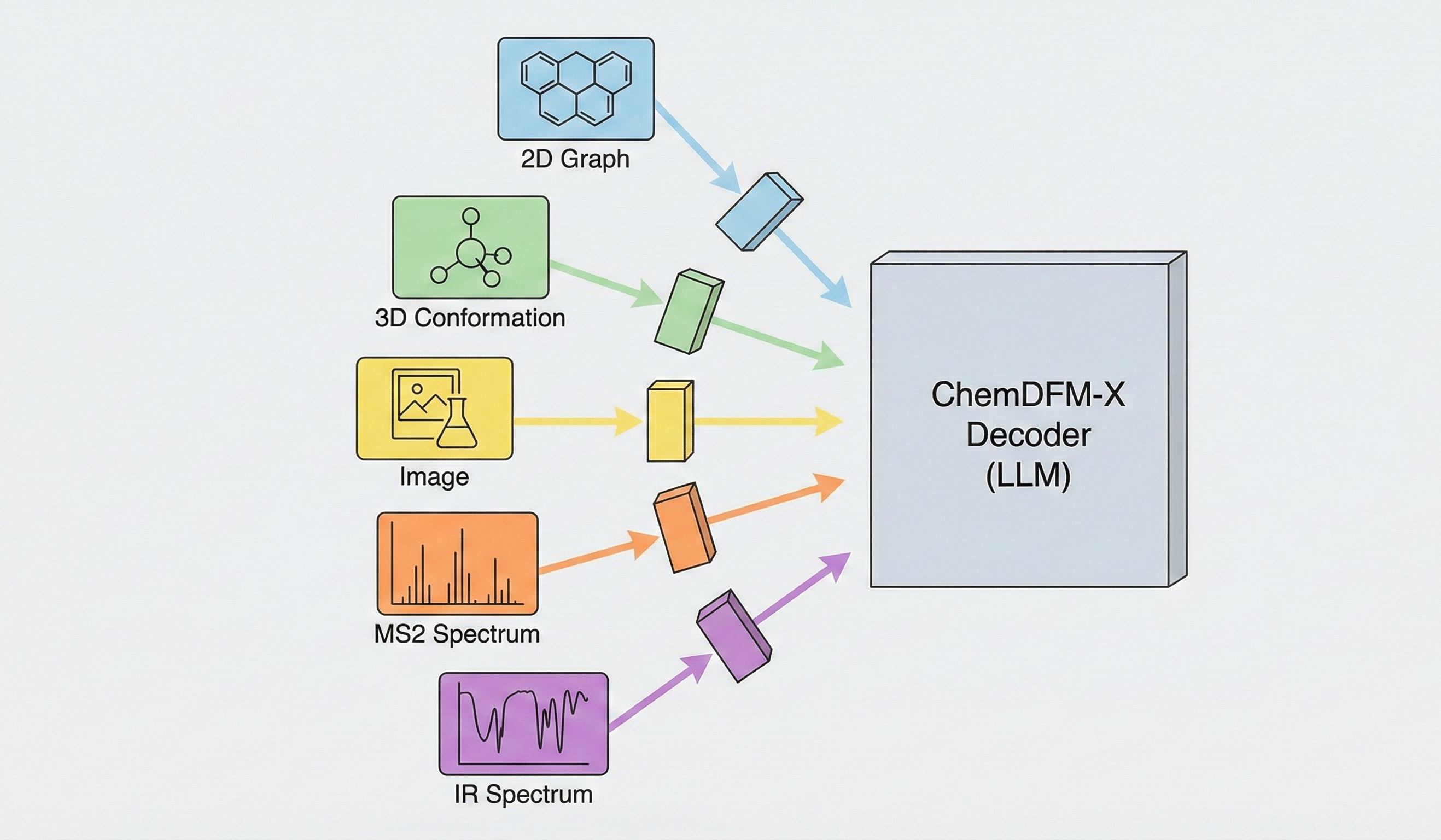
Multimodal chemical model integrating 5 modalities (2D graphs, 3D conformations, images, MS2/IR spectra) trained on 7.6M …
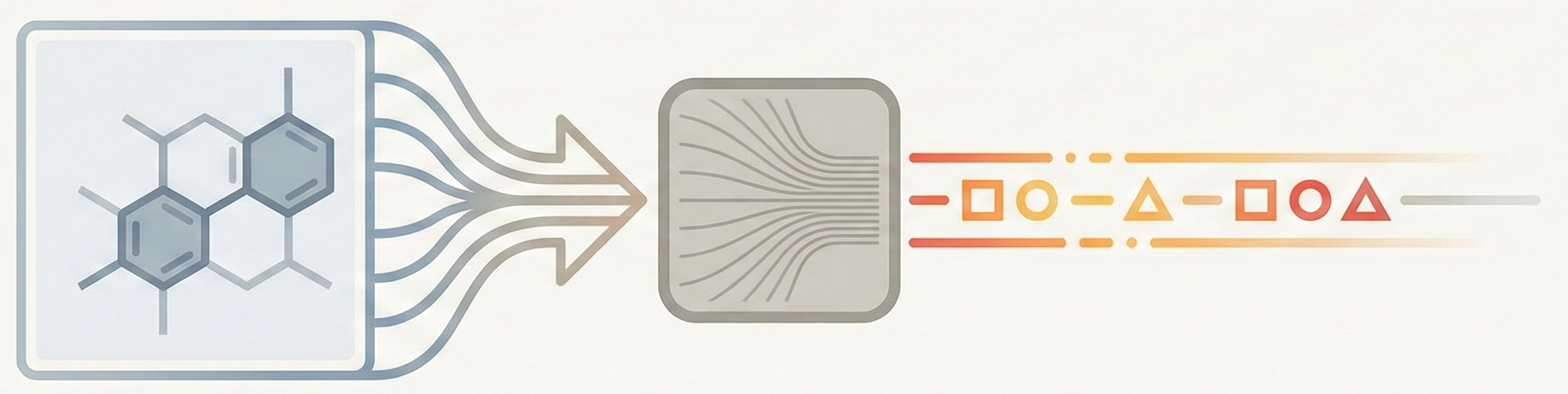
Comparative analysis of image-to-sequence OCSR methods across architecture, output format, training data, and compute …
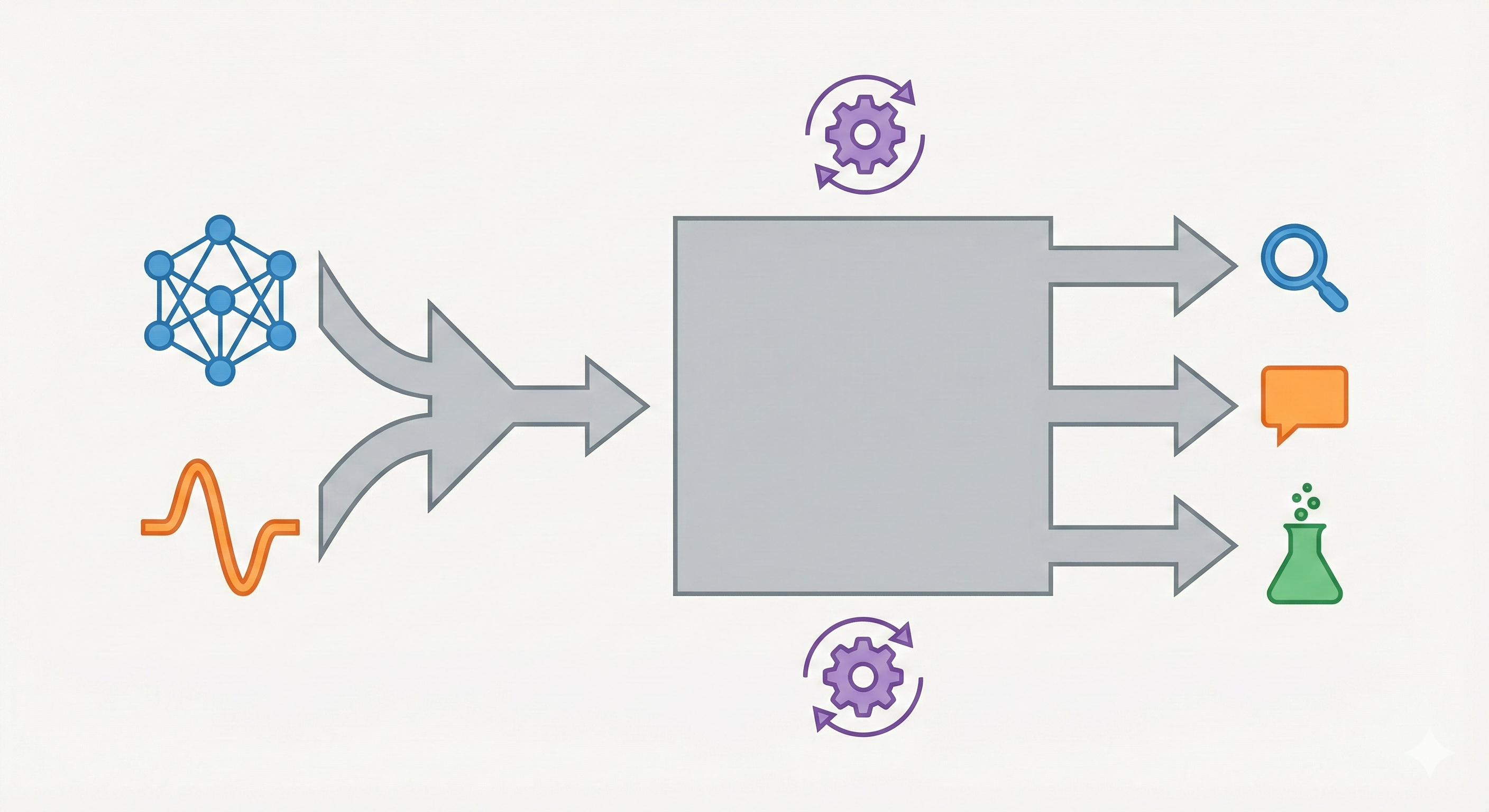
A multi-modal LLM aligning 2D molecular graphs with text via two-stage instruction tuning for drug discovery tasks.
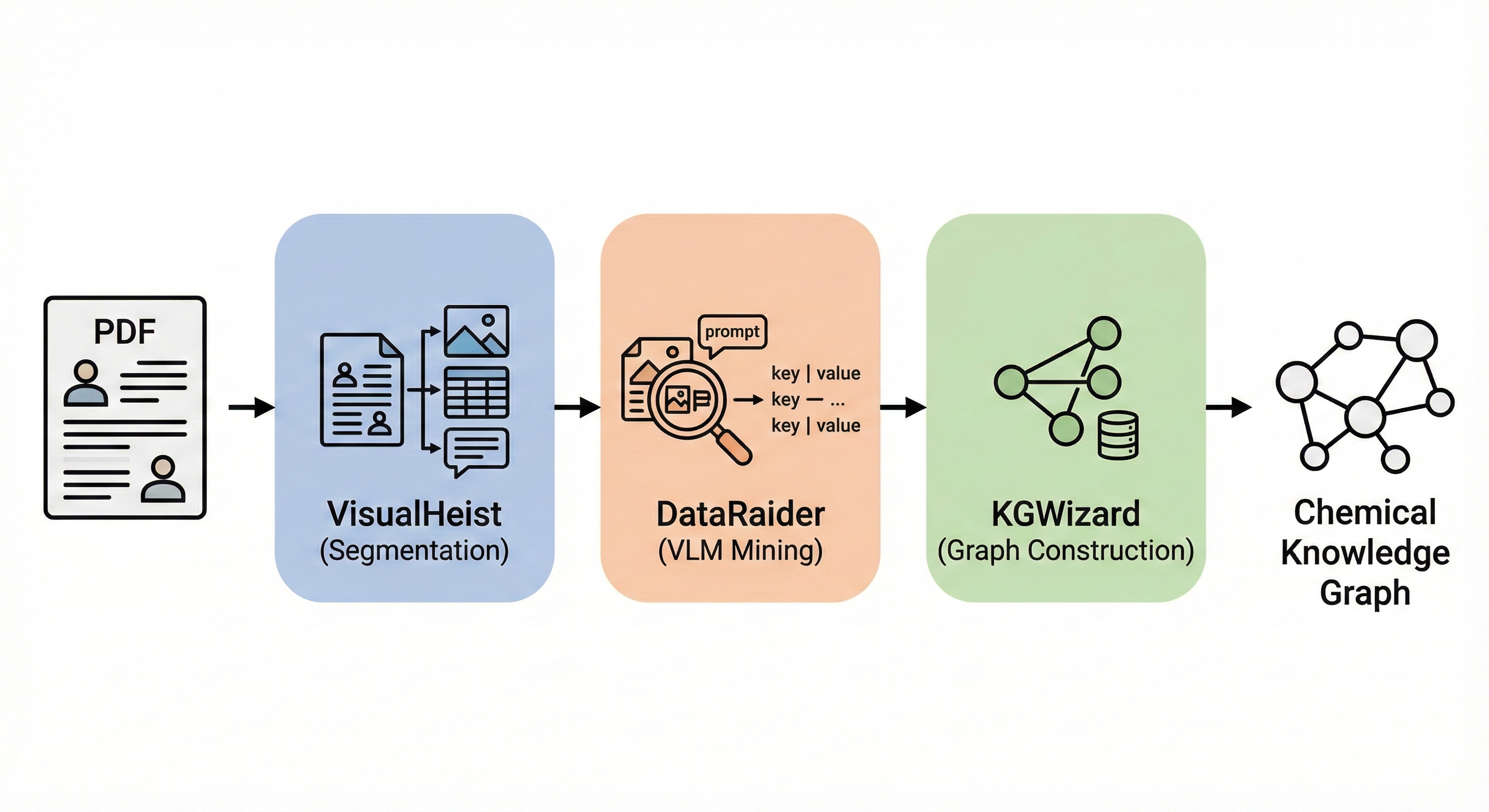
Vision-language pipeline extracting chemical reaction data from PDF figures and tables into structured knowledge graphs …
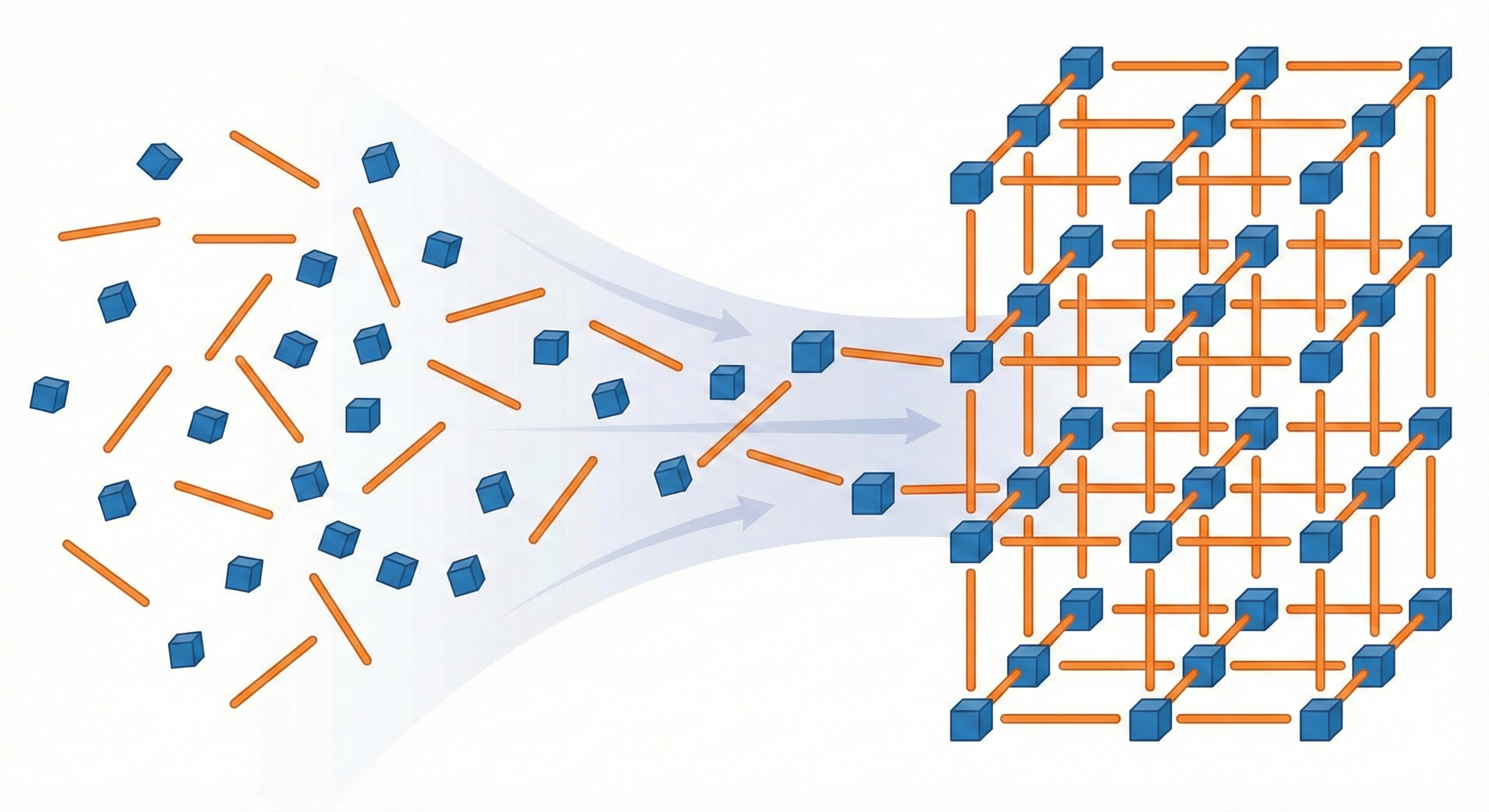
A Riemannian flow matching framework for generating Metal-Organic Framework structures by treating building blocks as …
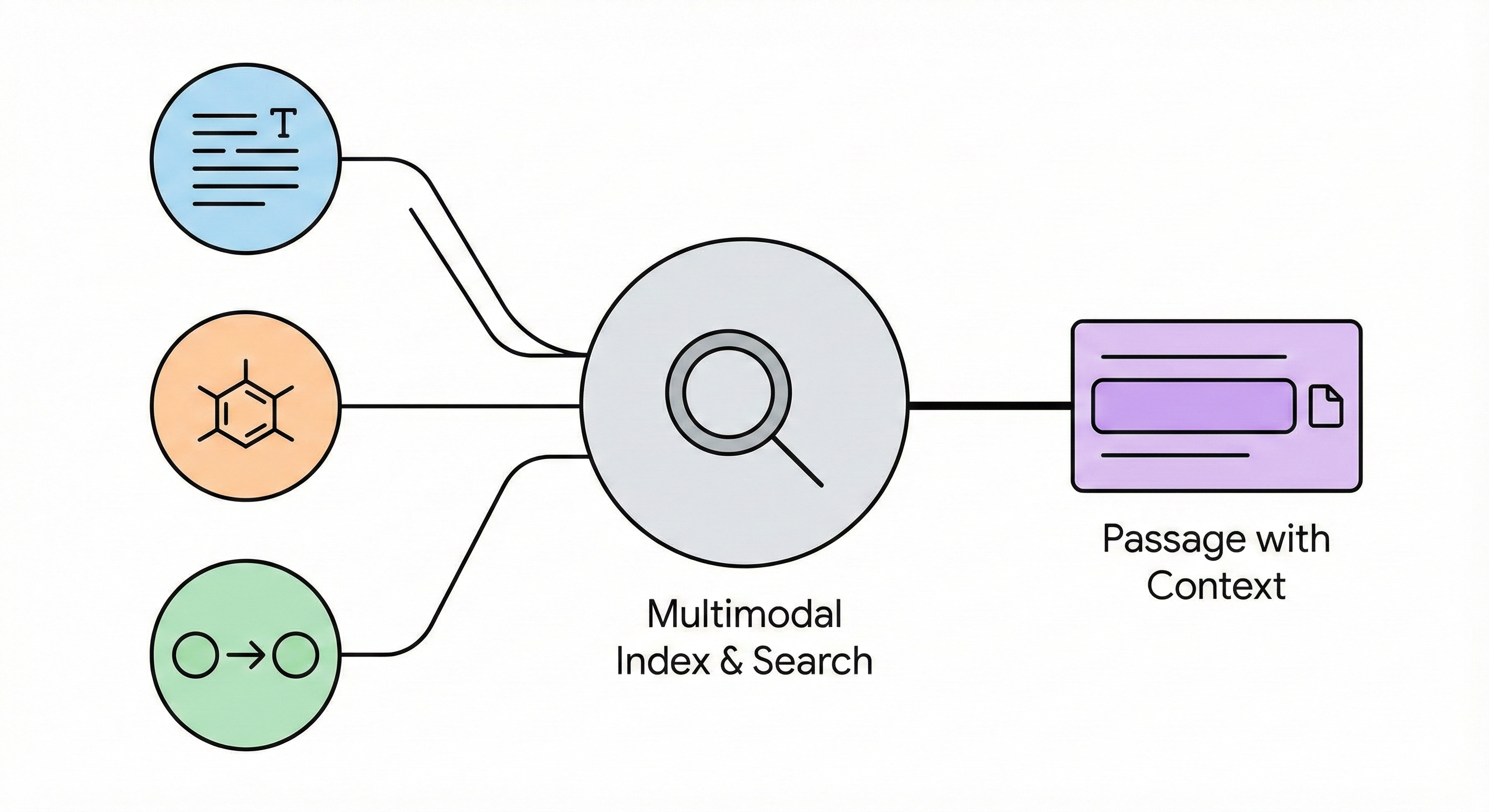
A multimodal search engine that integrates text passages, molecular diagrams, and reaction data to enable passage-level …