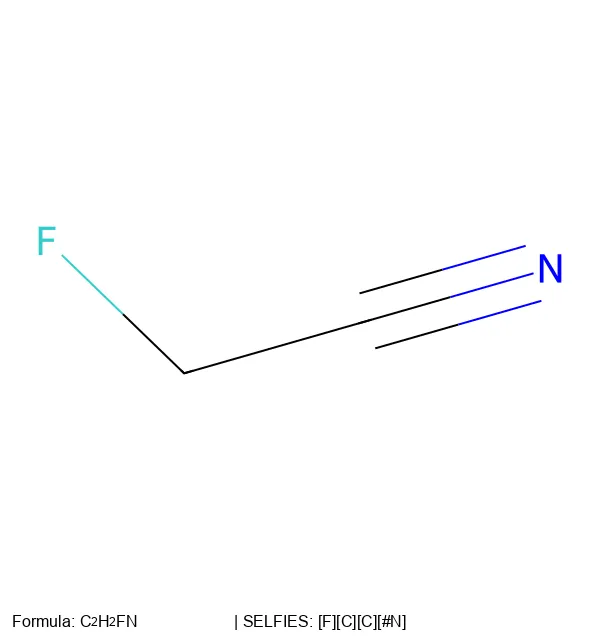
Converting SELFIES Strings to 2D Molecular Images
Create 2D molecular images from SELFIES strings using RDKit, SELFIES, and PIL, with proper formatting and legends.

Create 2D molecular images from SELFIES strings using RDKit, SELFIES, and PIL, with proper formatting and legends.
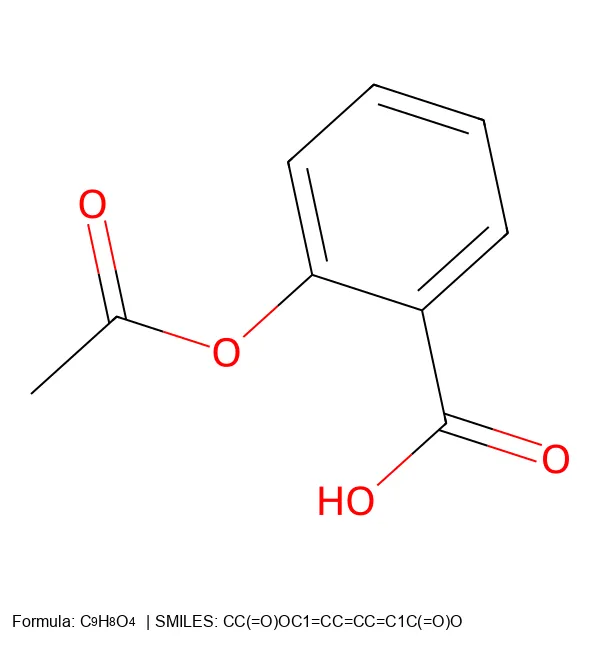
Create 2D molecular images from SMILES strings using RDKit and PIL, with proper formatting and legends.
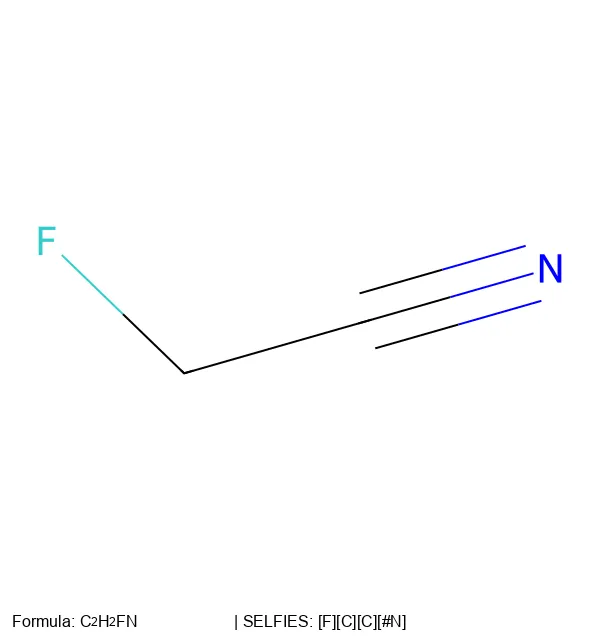
SELFIES is a 100% robust molecular string representation for ML, implemented in the open-source selfies Python library.
MARCEL dataset provides 722K+ conformers across 76K+ molecules for drug discovery, catalysis, and molecular …
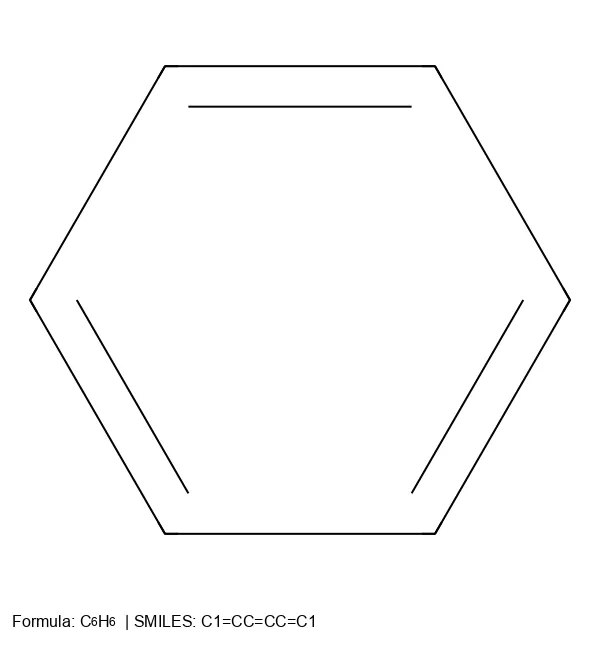
SMILES (Simplified Molecular Input Line Entry System) represents chemical structures using compact ASCII strings.
Dataset card for GEOM, providing energy-annotated molecular conformations generated via CREST/xTB and refined with DFT …
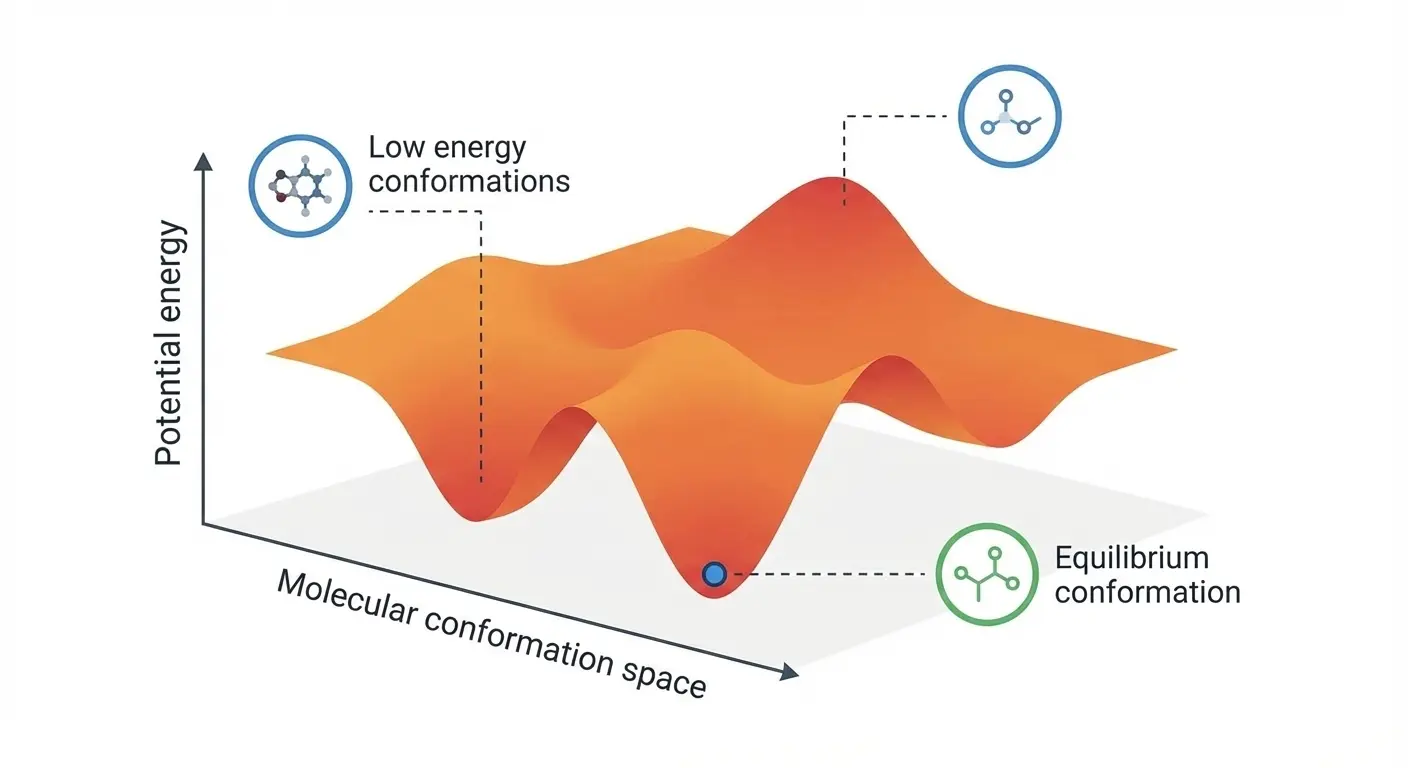
Liu et al.'s ICLR 2025 paper introducing DenoiseVAE, which learns adaptive, atom-specific noise for better molecular …
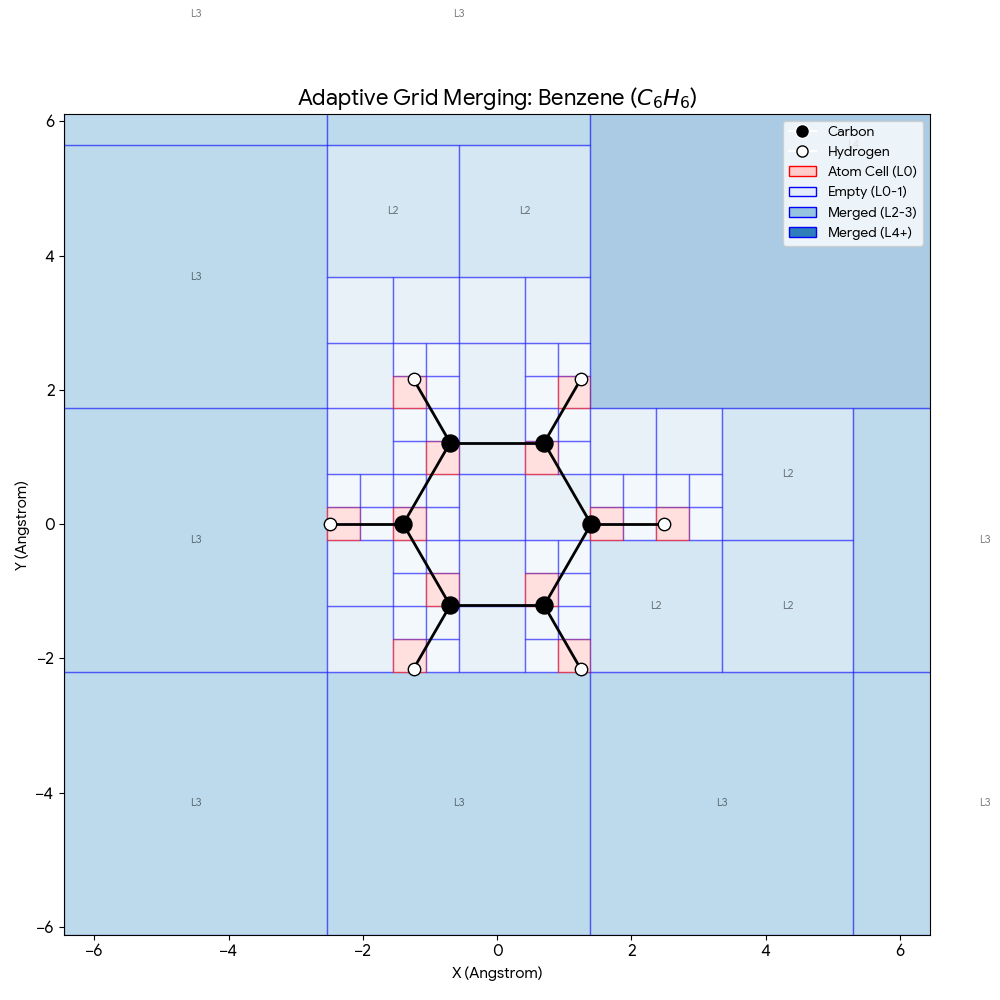
Lu et al. introduce SpaceFormer, a Transformer that models entire 3D molecular space (not just atoms) for superior …
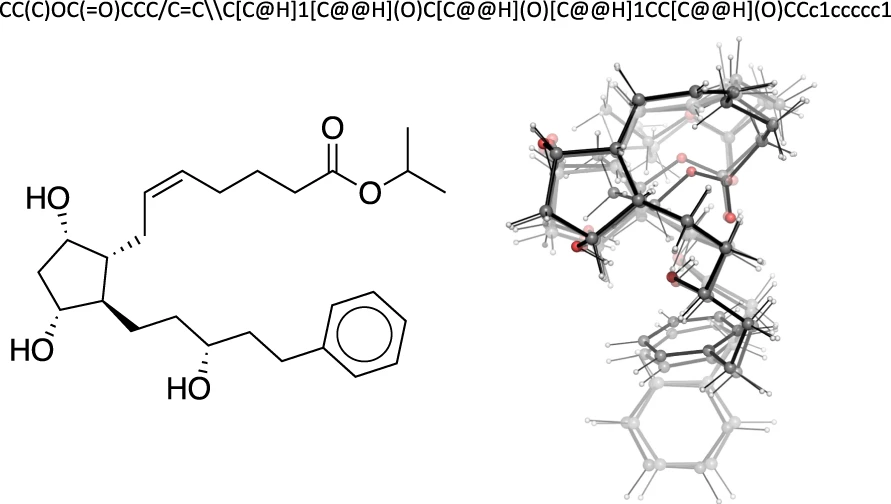
Learn how GEOM transforms 2D molecular graphs into dynamic 3D conformer ensembles for molecular machine learning …

Skinnider (2024) shows that generating invalid SMILES actually improves chemical language model performance through …
An end-to-end cheminformatics pipeline transforming 1D chemical formulas into 3D conformer datasets using graph …
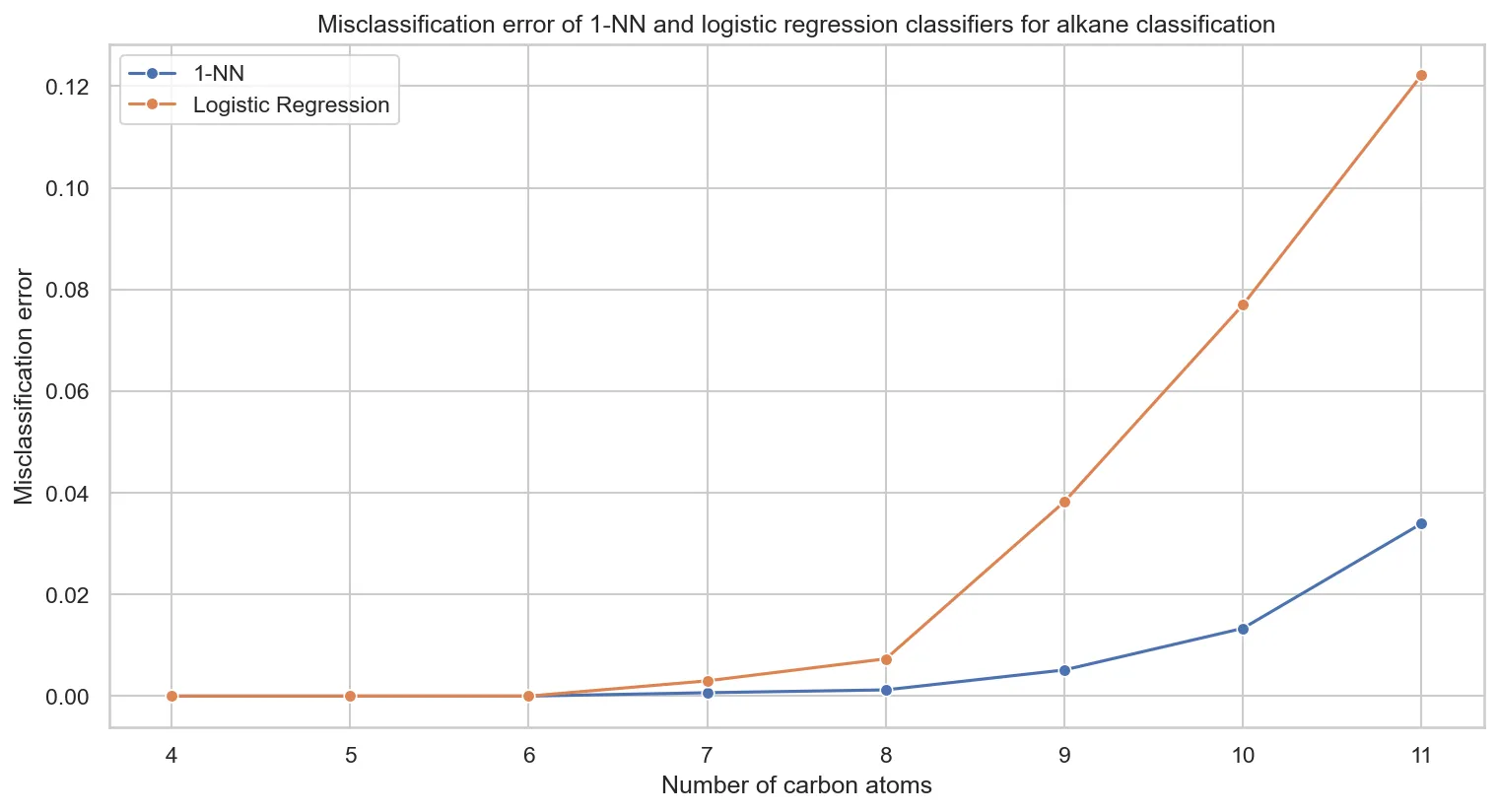
Supervised learning reveals hidden eigenvalue patterns that clustering missed, testing k-NN and logistic regression on …