
ChemBERTa-3: Open Source Training Framework
An open-source framework integrating DeepChem and Ray for training and benchmarking chemical foundation models like …

An open-source framework integrating DeepChem and Ray for training and benchmarking chemical foundation models like …
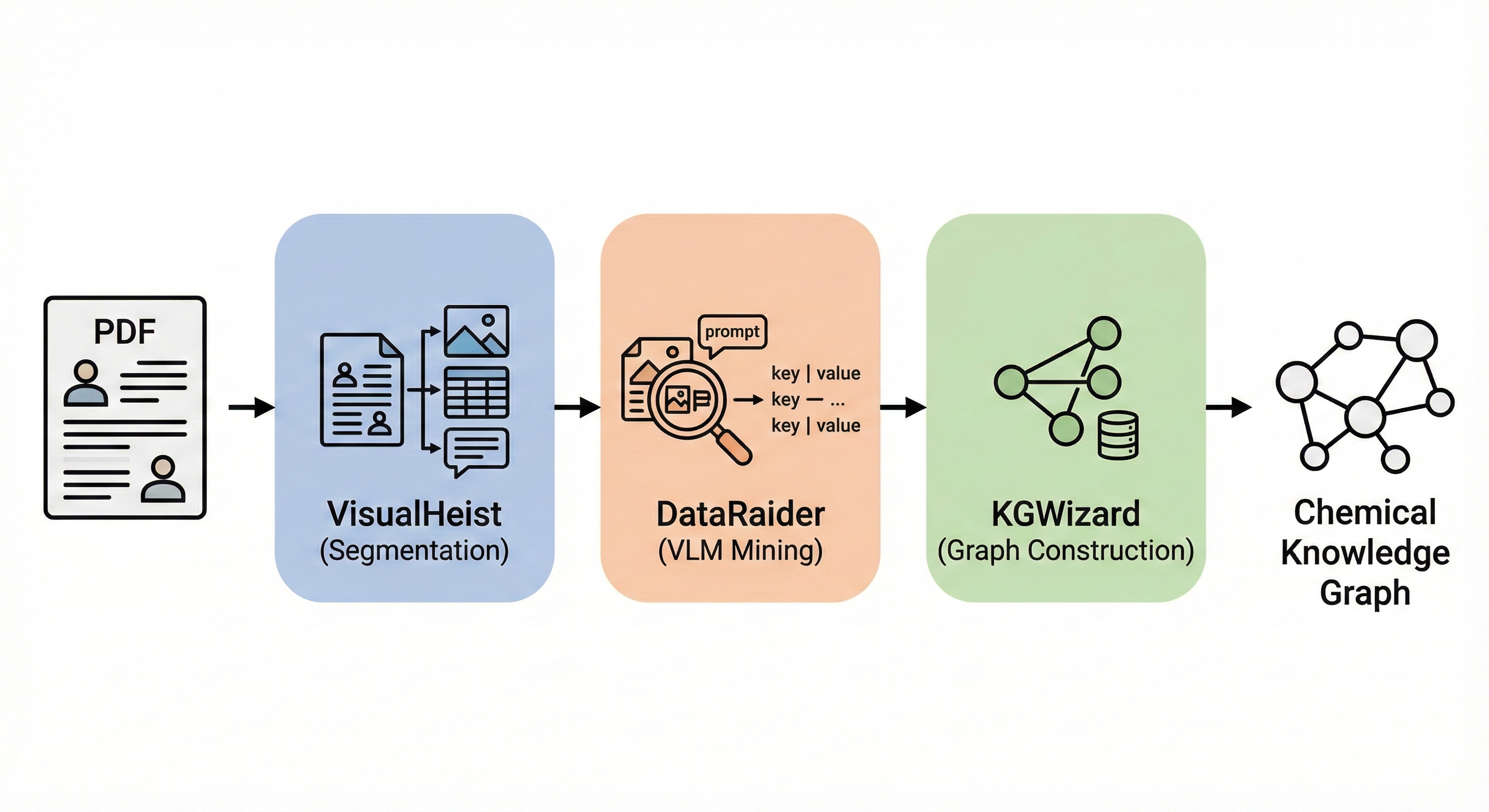
Vision-language pipeline extracting chemical reaction data from PDF figures and tables into structured knowledge graphs …
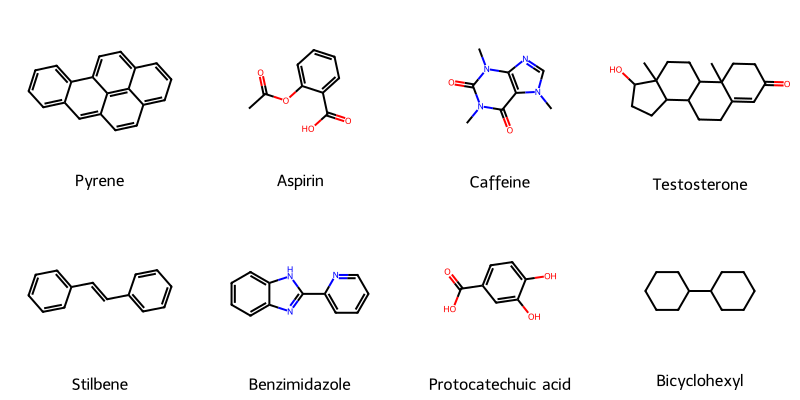
A robust, type-safe Python library for converting chemical strings (SMILES, SELFIES, InChI) into publication-quality …
Major updates to the SELFIES library, improved performance, expanded chemistry support, and new customization features.
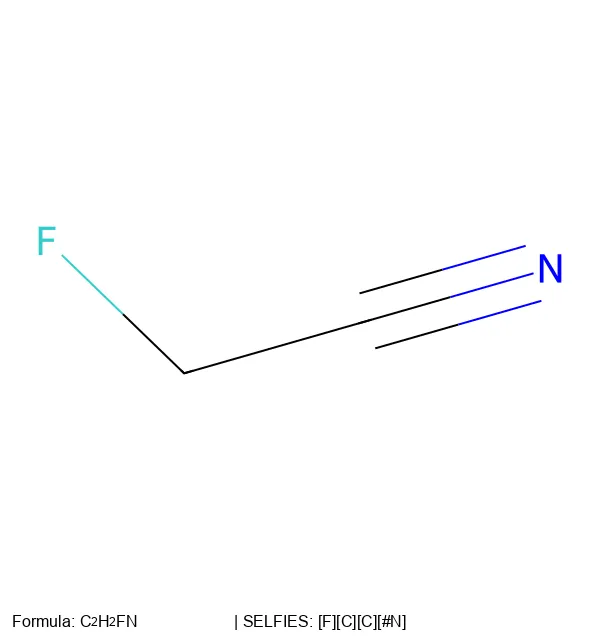
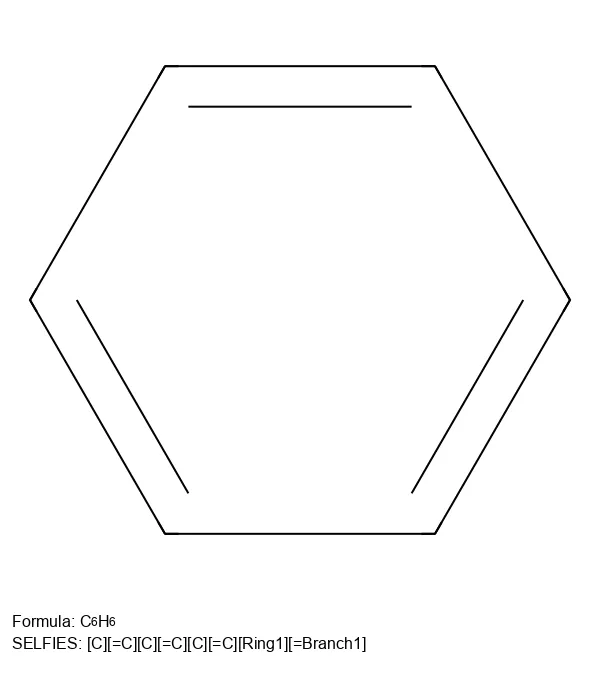
Create 2D molecular images from SELFIES strings using RDKit, SELFIES, and PIL, with proper formatting and legends.
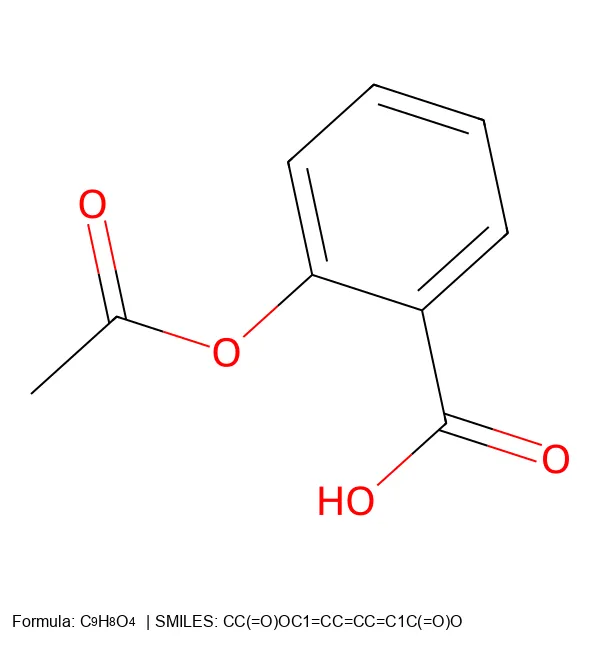
Create 2D molecular images from SMILES strings using RDKit and PIL, with proper formatting and legends.

Compare inverse transform sampling and von Neumann's rejection method for exponential random numbers with Python …
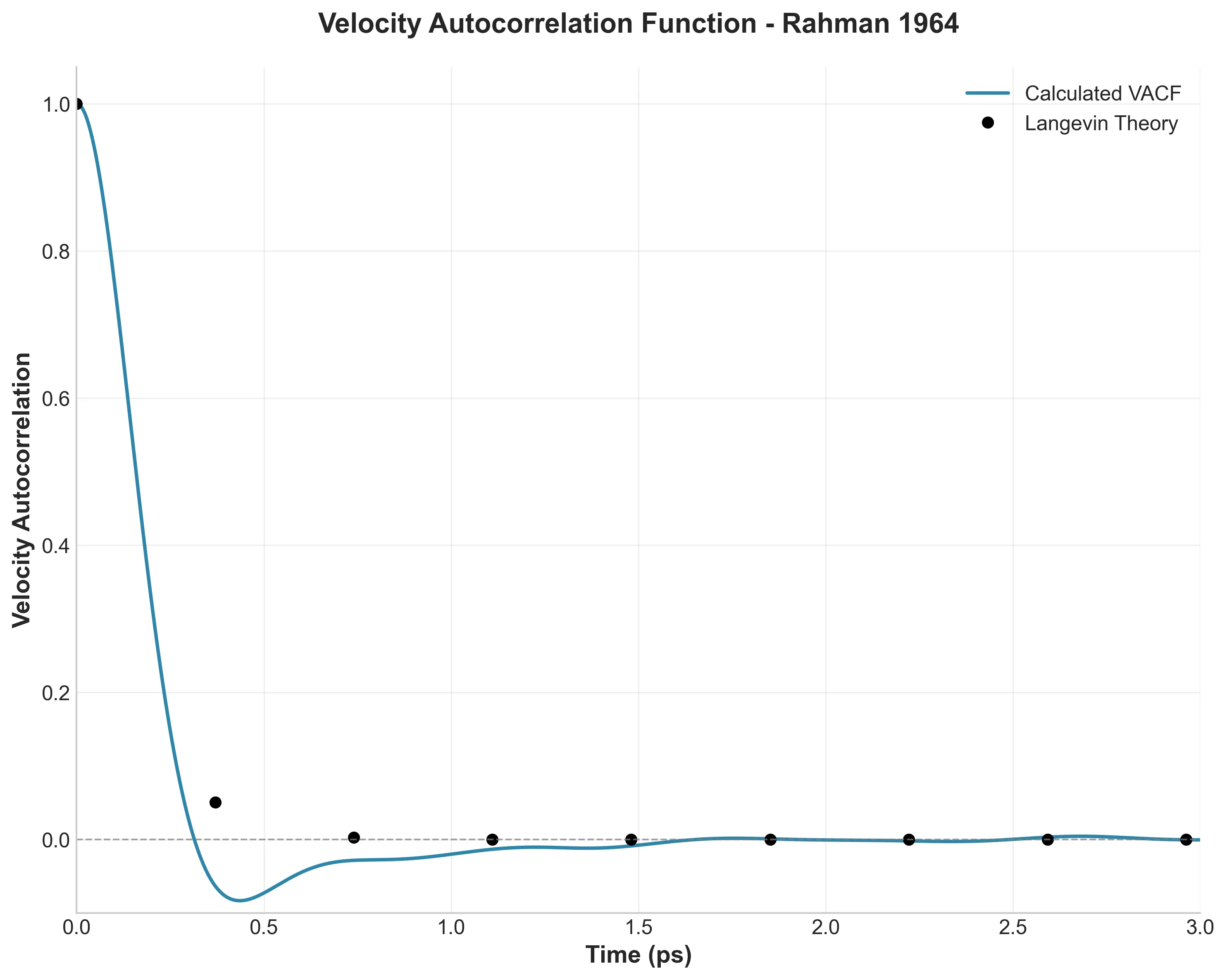
A high-fidelity replication of foundational molecular dynamics using modern software engineering practices: caching, …

How I used modern software engineering (caching, vectorization, and dependency locking) to reproduce a 60-year-old …
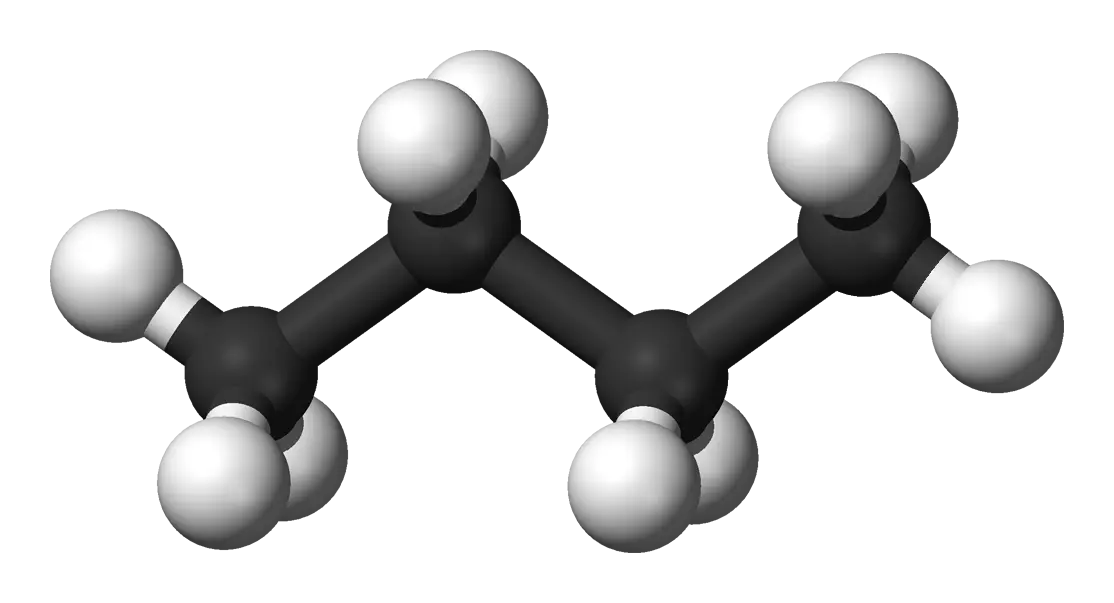
An end-to-end cheminformatics pipeline transforming 1D chemical formulas into 3D conformer datasets using graph …
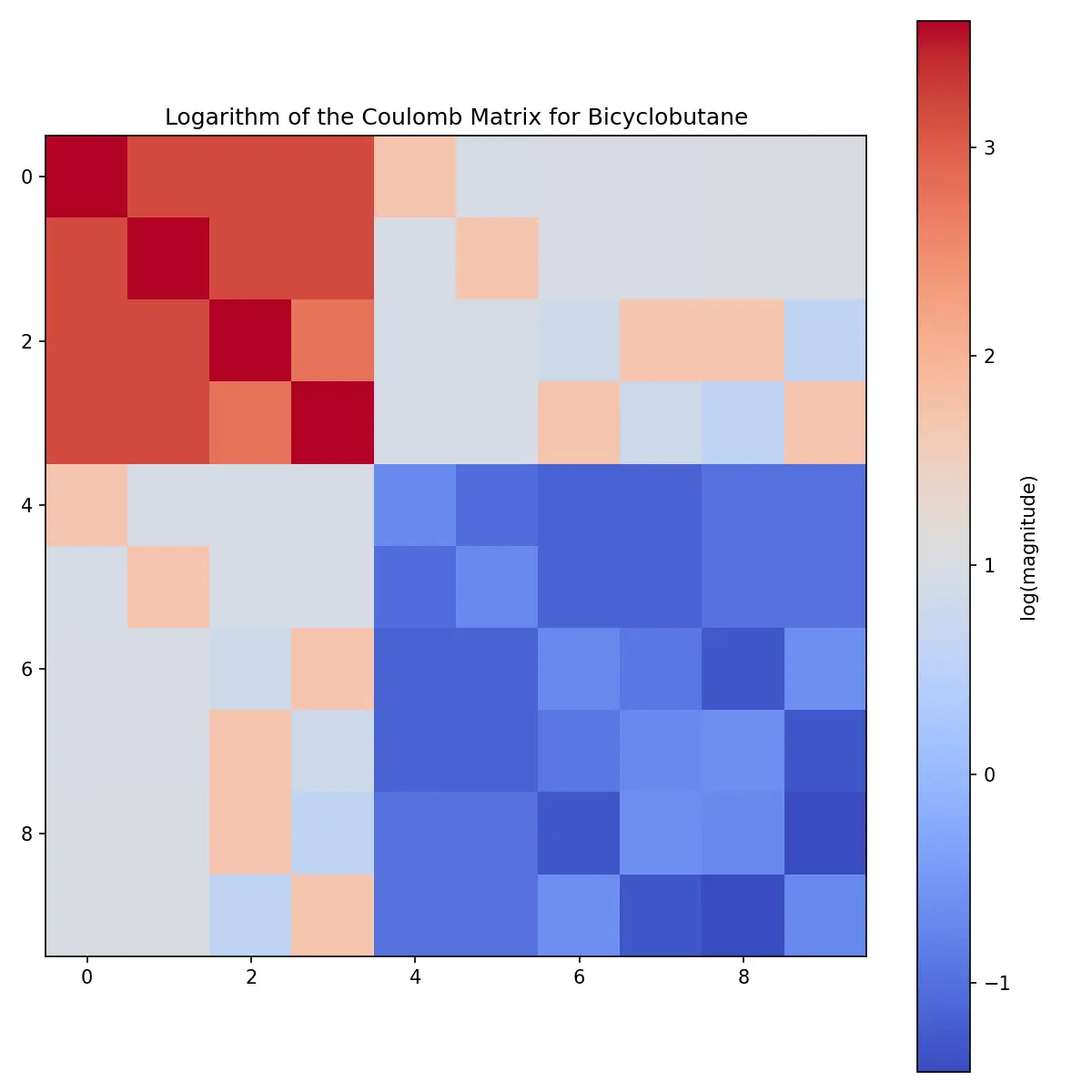
Learn how Coulomb matrices encode 3D molecular structure for machine learning from basic theory to Python implementation …
What happens to bills in Congress? Analyzing 15K+ bills from the 117th Congress to understand legislative patterns, …