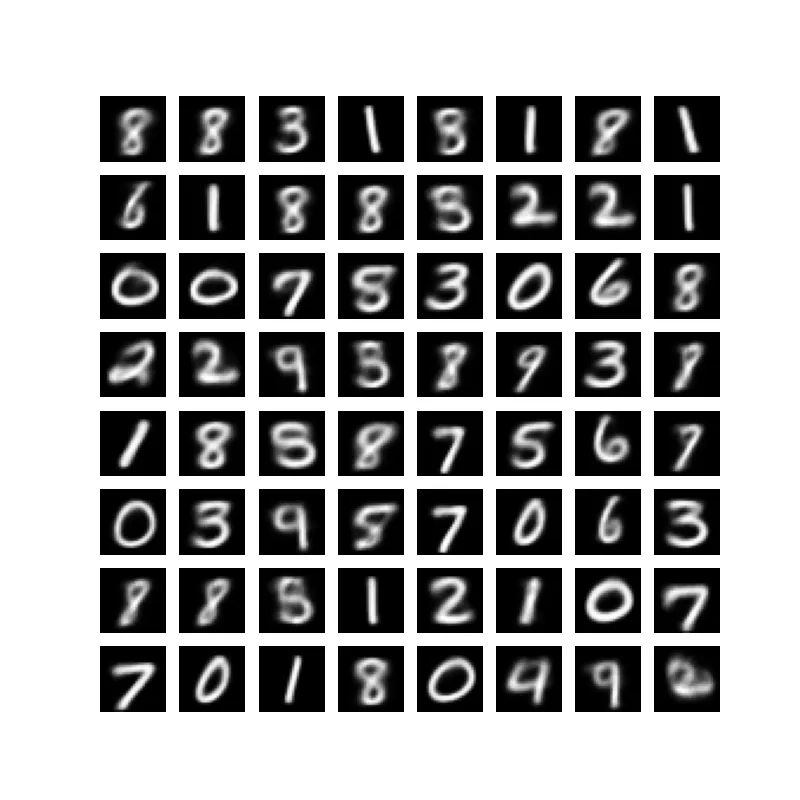
Importance Weighted Autoencoders: Beyond the Standard VAE
The key difference between multi-sample VAEs and IWAEs: how log-of-averages creates a tighter bound on log-likelihood.

The key difference between multi-sample VAEs and IWAEs: how log-of-averages creates a tighter bound on log-likelihood.

Guide to implementing the Müller-Brown potential in PyTorch, comparing analytical vs automatic differentiation with …

Langevin dynamics simulation showing particle motion in the deep reactant minimum (Basin MA) of the Müller-Brown …

Langevin dynamics simulation showing particle motion in the product minimum (Basin MB) of the Müller-Brown potential …

GPU-accelerated PyTorch framework for the Müller-Brown potential with JIT compilation, Langevin dynamics, and …

Extended Langevin dynamics simulation showing particle transitions between different basins of the Müller-Brown …
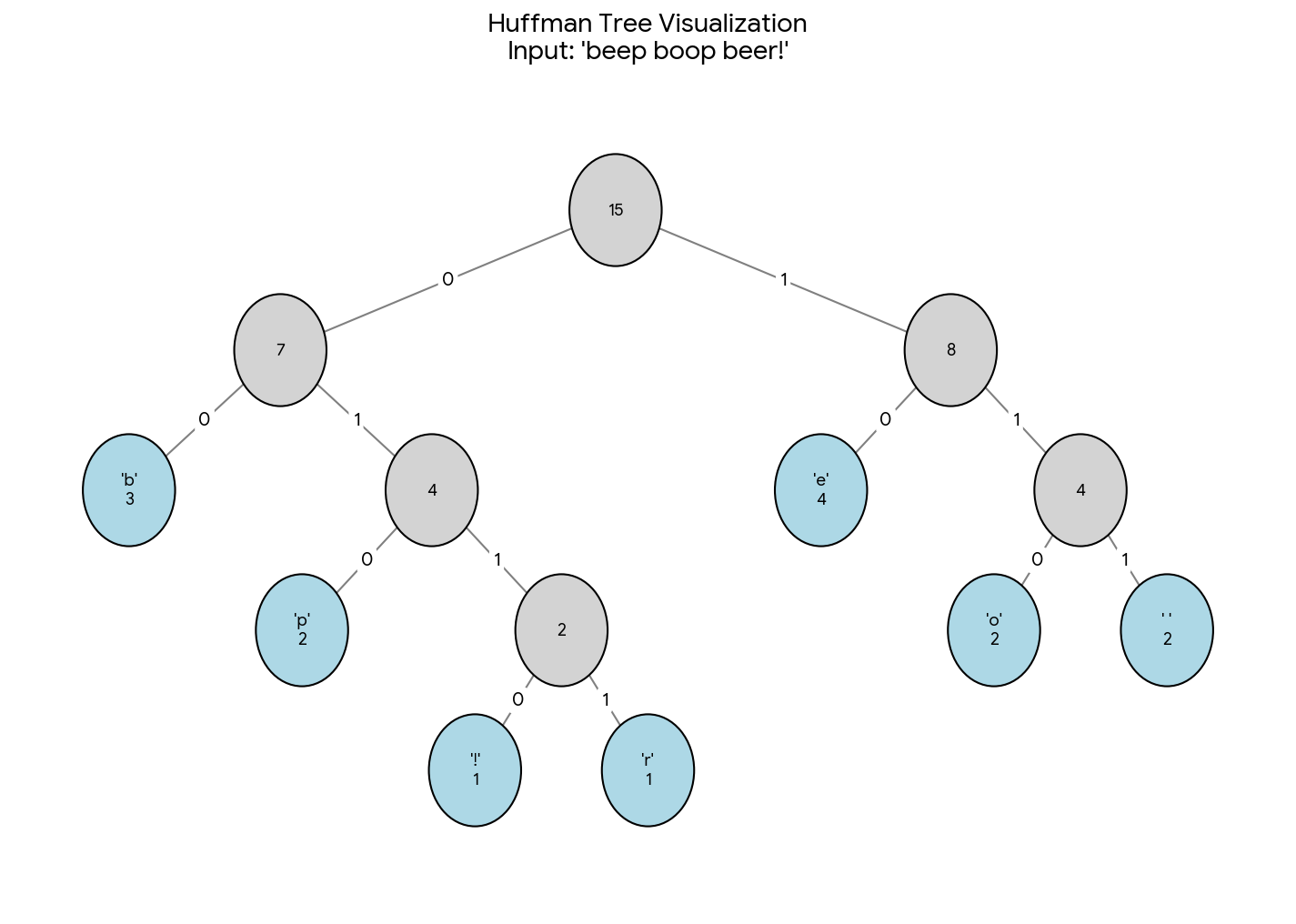
Production-grade Word2Vec in PyTorch with vectorized Hierarchical Softmax, Negative Sampling, and torch.compile support.
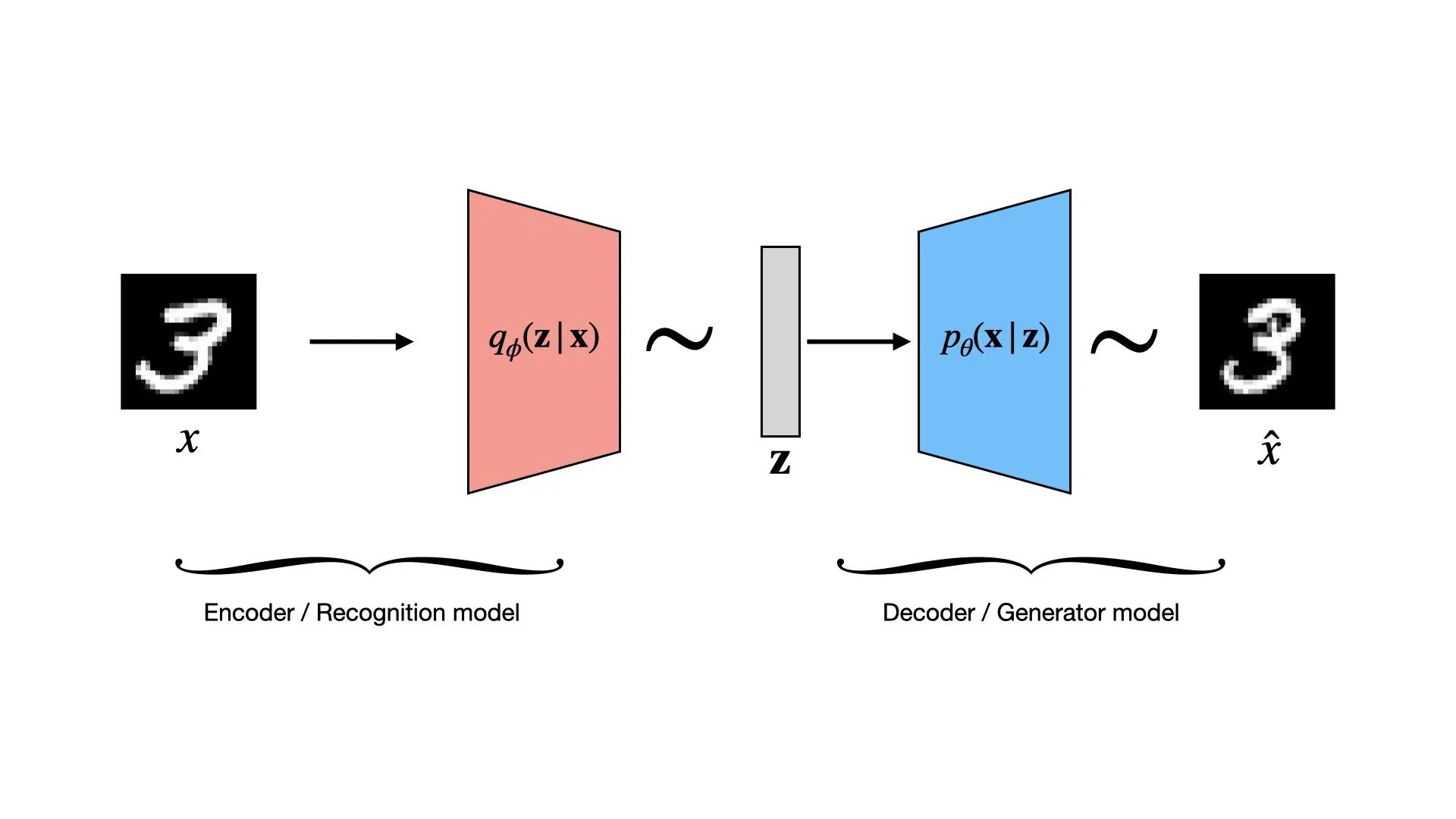
Learn to implement VAEs in PyTorch: ELBO objective, reparameterization trick, loss scaling, and MNIST experiments on …

Learn about the Kabsch algorithm for optimal point alignment with implementations in NumPy, PyTorch, TensorFlow, and JAX …
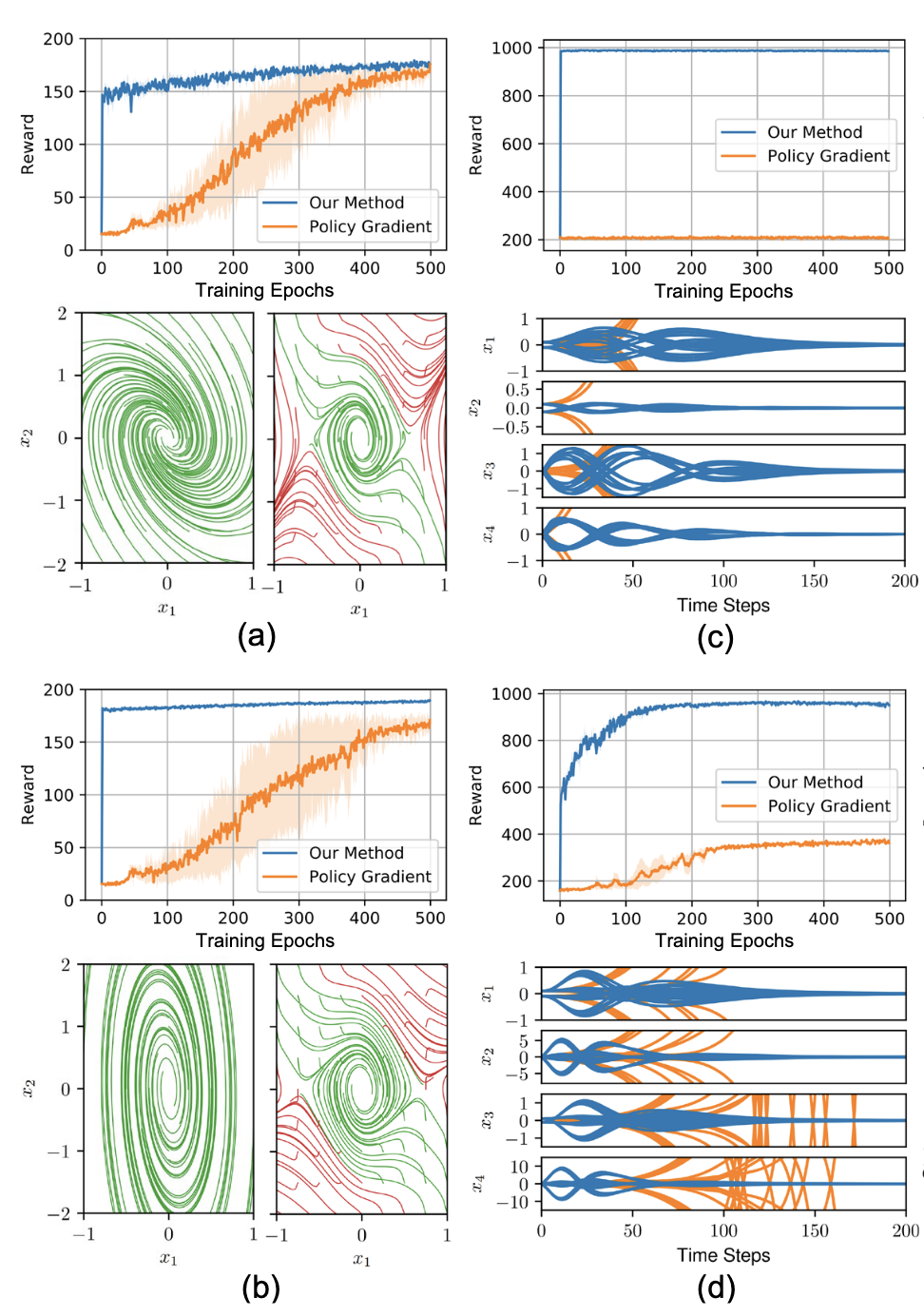
PyTorch IQCRNN enforcing stability guarantees on RNNs via Integral Quadratic Constraints and semidefinite programming.